Cortical Surface Reconstruction from High-Resolution MR Brain Images
- PMID: 22481909
- PMCID: PMC3296314
- DOI: 10.1155/2012/870196
Cortical Surface Reconstruction from High-Resolution MR Brain Images
Abstract
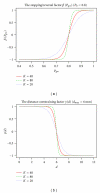
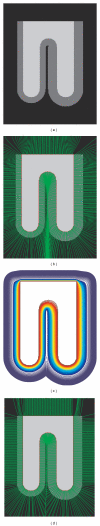
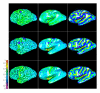
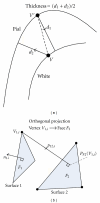
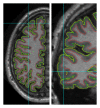
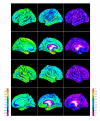
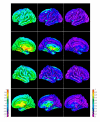
Reconstruction of the cerebral cortex from magnetic resonance (MR) images is an important step in quantitative analysis of the human brain structure, for example, in sulcal morphometry and in studies of cortical thickness. Existing cortical reconstruction approaches are typically optimized for standard resolution (~1 mm) data and are not directly applicable to higher resolution images. A new PDE-based method is presented for the automated cortical reconstruction that is computationally efficient and scales well with grid resolution, and thus is particularly suitable for high-resolution MR images with submillimeter voxel size. The method uses a mathematical model of a field in an inhomogeneous dielectric. This field mapping, similarly to a Laplacian mapping, has nice laminar properties in the cortical layer, and helps to identify the unresolved boundaries between cortical banks in narrow sulci. The pial cortical surface is reconstructed by advection along the field gradient as a geometric deformable model constrained by topology-preserving level set approach. The method's performance is illustrated on exvivo images with 0.25-0.35 mm isotropic voxels. The method is further evaluated by cross-comparison with results of the FreeSurfer software on standard resolution data sets from the OASIS database featuring pairs of repeated scans for 20 healthy young subjects.
Figures





References
-
- Han X, Pham DL, Tosun D, Rettmann ME, Xu C, Prince JL. CRUISE: cortical reconstruction using implicit surface evolution. NeuroImage. 2004;23(3):997–1012. - PubMed
-
- Kim JS, Singh V, Lee JK, et al. Automated 3-D extraction and evaluation of the inner and outer cortical surfaces using a Laplacian map and partial volume effect classification. NeuroImage. 2005;27(1):210–221. - PubMed
-
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis: i. segmentation and surface reconstruction. NeuroImage. 1999;9(2):179–194. - PubMed
-
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis: II. Inflation, flattening, and a surface-based coordinate system. NeuroImage. 1999;9(2):195–207. - PubMed
-
- Fischl B, Liu A, Dale A. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Transactions on Medical Imaging. 2001;20(1):70–80. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

