In silico characterization of a nitrate reductase gene family and analysis of the predicted proteins from the moss Physcomitrella patens
- PMID: 22482004
- PMCID: PMC3291307
- DOI: 10.4161/cib.18534
In silico characterization of a nitrate reductase gene family and analysis of the predicted proteins from the moss Physcomitrella patens
Abstract
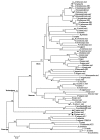
Assimilatory nitrate reductase (NR; EC 1.7.1.1-3) catalyzes the reduction of nitrate to nitrite. This enzyme has a conserved structure common to fungi, algae and plants. However, some differences in the amino acid sequence between plant and algal NR suggest that the activity regulation mechanisms have changed during plant evolution. Since only NRs from angiosperms have been studied, the search and analysis of NR genes and proteins from the moss Physcomitrella patens, a basal land plant, was performed to widen the knowledge of land plant NR structure. A family of three nr genes, named ppnia1;1, ppnia1;2 and ppnia2, was localized in the P. patens genome. The predicted proteins are canonical NRs with the conserved domains Molybdene-Cytochorme b -Cytochrome b reductase and possess 20 amino acid residues important for the enzymatic function conserved in plant and algal NRs. Interestingly, moss NRs lack a consensus sequence, common to angiosperm NRs, that is a target for posttranslational regulation. A phylogenetic tree with embryophyte and green algae NR sequences was constructed and P. patens NRs localized at the base of embryophyte NR evolution. The data presented here suggest that bryophytes and vascular plants have different systems to regulate NR activity.
Keywords: Physcomitrella patens; basal embryophytes; nitrate reductase; posttranslational regulation.
Figures
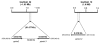

Similar articles
-
The nitric oxide production in the moss Physcomitrella patens is mediated by nitrate reductase.PLoS One. 2015 Mar 5;10(3):e0119400. doi: 10.1371/journal.pone.0119400. eCollection 2015. PLoS One. 2015. PMID: 25742644 Free PMC article.
-
Post-translational control of nitrate reductase activity responding to light and photosynthesis evolved already in the early vascular plants.J Plant Physiol. 2013 May 1;170(7):662-7. doi: 10.1016/j.jplph.2012.12.010. Epub 2013 Feb 8. J Plant Physiol. 2013. PMID: 23395536
-
The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants.Mol Biol Evol. 2007 Mar;24(3):699-709. doi: 10.1093/molbev/msl198. Epub 2006 Dec 14. Mol Biol Evol. 2007. PMID: 17175527
-
Adaptation Mechanisms in the Evolution of Moss Defenses to Microbes.Front Plant Sci. 2017 Mar 15;8:366. doi: 10.3389/fpls.2017.00366. eCollection 2017. Front Plant Sci. 2017. PMID: 28360923 Free PMC article. Review.
-
Light-harvesting antenna complexes in the moss Physcomitrella patens: implications for the evolutionary transition from green algae to land plants.Curr Opin Plant Biol. 2017 Jun;37:94-101. doi: 10.1016/j.pbi.2017.04.002. Epub 2017 Apr 23. Curr Opin Plant Biol. 2017. PMID: 28445834 Review.
Cited by
-
Fast-track transformation and genome editing in Brachypodium distachyon.Plant Methods. 2023 Mar 29;19(1):31. doi: 10.1186/s13007-023-01005-1. Plant Methods. 2023. PMID: 36991448 Free PMC article.
-
Understanding nitrate assimilation and its regulation in microalgae.Front Plant Sci. 2015 Oct 26;6:899. doi: 10.3389/fpls.2015.00899. eCollection 2015. Front Plant Sci. 2015. PMID: 26579149 Free PMC article. Review.
-
Expression of novel nitrate reductase genes in the harmful alga, Chattonella subsalsa.Sci Rep. 2018 Sep 7;8(1):13417. doi: 10.1038/s41598-018-31735-5. Sci Rep. 2018. PMID: 30194416 Free PMC article.
-
The nitric oxide production in the moss Physcomitrella patens is mediated by nitrate reductase.PLoS One. 2015 Mar 5;10(3):e0119400. doi: 10.1371/journal.pone.0119400. eCollection 2015. PLoS One. 2015. PMID: 25742644 Free PMC article.
References
LinkOut - more resources
Full Text Sources
