TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling
- PMID: 22482711
- PMCID: PMC3322363
- DOI: 10.1016/j.coviro.2012.02.003
TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling
Abstract
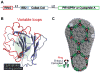
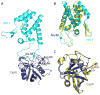
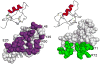
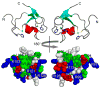
TRIM5 is a restriction factor that blocks retrovirus infection soon after the virion core enters the cell cytoplasm. Restriction activity is targeted to the virion core via recognition of the capsid protein lattice that encases the viral genomic RNA. In common with all of the many TRIM family members, TRIM5 has RING, B-box, and coiled-coil domains. As an E3 ubiquitin ligase TRIM5 cooperates with the heterodimeric E2, UBC13/UEV1A, to activate the TAK1 (MAP3K7) kinase, NF-κB and AP-1 signaling, and the transcription of inflammatory cytokines and chemokines. TAK1, UBC13, and UEV1A all contribute to TRIM5-mediated retrovirus restriction activity. Interaction of the carboxy-terminal PRYSPRY or cyclophilin domains of TRIM5 with the retroviral capsid lattice stimulates the formation of a complementary lattice by TRIM5, with greatly increased TRIM5 E3 activity, and host cell signal transduction. Structural and biochemical studies on TRIM5 have opened a much needed window on how the innate immune system detects the distinct molecular features of HIV-1 and other retroviruses.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures

References
-
- Nisole S, Stoye JP, Saïb A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

