Pentachlorophenol hydroxylase, a poorly functioning enzyme required for degradation of pentachlorophenol by Sphingobium chlorophenolicum
- PMID: 22482720
- PMCID: PMC4251658
- DOI: 10.1021/bi300261p
Pentachlorophenol hydroxylase, a poorly functioning enzyme required for degradation of pentachlorophenol by Sphingobium chlorophenolicum
Abstract
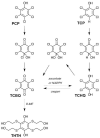
Several strains of Sphingobium chlorophenolicum have been isolated from soil that was heavily contaminated with pentachlorophenol (PCP), a toxic pesticide introduced in the 1930s. S. chlorophenolicum appears to have assembled a poorly functioning pathway for degradation of PCP by patching enzymes recruited via two independent horizontal gene transfer events into an existing metabolic pathway. Flux through the pathway is limited by PCP hydroxylase. PCP hydroxylase is a dimeric protein that belongs to the family of flavin-dependent phenol hydroxylases. In the presence of NADPH, PCP hydroxylase converts PCP to tetrachlorobenzoquinone (TCBQ). The k(cat) for PCP (0.024 s(-1)) is very low, suggesting that the enzyme is not well evolved for turnover of this substrate. Structure-activity studies reveal that substrate binding and activity are enhanced by a low pK(a) for the phenolic proton, increased hydrophobicity, and the presence of a substituent ortho to the hydroxyl group of the phenol. PCP hydroxylase exhibits substantial uncoupling; the C4a-hydroxyflavin intermediate, instead of hydroxylating the substrate, can decompose to produce H(2)O(2) in a futile cycle that consumes NADPH. The extent of uncoupling varies from 0 to 100% with different substrates. The extent of uncoupling is increased by the presence of bulky substituents at position 3, 4, or 5 and decreased by the presence of a chlorine in the ortho position. The effectiveness of PCP hydroxylase is additionally hindered by its promiscuous activity with tetrachlorohydroquinone (TCHQ), a downstream metabolite in the degradation pathway. The conversion of TCHQ to TCBQ reverses flux through the pathway. Substantial uncoupling also occurs during the reaction with TCHQ.
Figures





References
-
- Miller LL, Ingerman LD, Singh M. Toxicological profile for pentachlorophenol. Agency for Toxic Substances and Disease Registry, U.S. Department of Health and Human Services; 2001.
-
- Lin P-H, Nakamura J, Yamaguchi S, Upton PB, La DK, Swenberg JA. Oxidative damage and direct adducts in calf thymus DNA induced by the pentachlorophenol metabolites, tetrachlorohydroquinone and tetrachloro-1,4-benzoquinone. Carcinogenesis. 2001;22:627–634. - PubMed
-
- Weinbach EC. The effect of pentachlorophenol on oxidative phosphorylation. J Biol Chem. 1954;210:545–550. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information
Molecular Biology Databases
Miscellaneous

