Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal
- PMID: 22482728
- PMCID: PMC4844853
- DOI: 10.1016/j.cmet.2012.02.013
Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal
Abstract
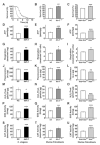
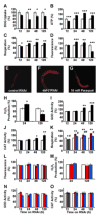
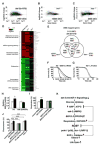
Impaired insulin and IGF-1 signaling (iIIS) in C. elegans daf-2 mutants extends life span more than 2-fold. Constitutively, iIIS increases mitochondrial activity and reduces reactive oxygen species (ROS) levels. By contrast, acute impairment of daf-2 in adult C. elegans reduces glucose uptake and transiently increases ROS. Consistent with the concept of mitohormesis, this ROS signal causes an adaptive response by inducing ROS defense enzymes (SOD, catalase), culminating in ultimately reduced ROS levels despite increased mitochondrial activity. Inhibition of this ROS signal by antioxidants reduces iIIS-mediated longevity by up to 60%. Induction of the ROS signal requires AAK-2 (AMPK), while PMK-1 (p38) and SKN-1 (NRF-2) are needed for the retrograde response. IIIS upregulates mitochondrial L-proline catabolism, and impairment of the latter impairs the life span-extending capacity of iIIS while L-proline supplementation extends C. elegans life span. Taken together, iIIS promotes L-proline metabolism to generate a ROS signal for the adaptive induction of endogenous stress defense to extend life span.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Alternative mitochondrial fuel extends life span.Cell Metab. 2012 Apr 4;15(4):417-8. doi: 10.1016/j.cmet.2012.03.011. Cell Metab. 2012. PMID: 22482723 Free PMC article.
References
-
- Al-Regaiey KA, Masternak MM, Bonkowski M, Sun L, Bartke A. Long-lived growth hormone receptor knockout mice: interaction of reduced insulin-like growth factor i/insulin signaling and caloric restriction. Endocrinology. 2005;146:851–860. - PubMed
-
- Barros MH, Bandy B, Tahara EB, Kowaltowski AJ. Higher respiratory activity decreases mitochondrial reactive oxygen release and increases life span in Saccharomyces cerevisiae. J Biol Chem. 2004;279:49883–49888. - PubMed
-
- Bartke A, Wright JC, Mattison JA, Ingram DK, Miller RA, Roth GS. Extending the lifespan of long-lived mice. Nature. 2001;414:412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

