The long and the short of it: the role of the zinc finger polyadenosine RNA binding protein, Nab2, in control of poly(A) tail length
- PMID: 22484098
- PMCID: PMC3345082
- DOI: 10.1016/j.bbagrm.2012.03.006
The long and the short of it: the role of the zinc finger polyadenosine RNA binding protein, Nab2, in control of poly(A) tail length
Abstract
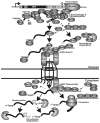
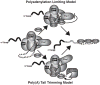
In eukaryotic cells, addition of poly(A) tails to transcripts by 3'-end processing/polyadenylation machinery is a critical step in gene expression. The length of the poly(A) tail influences the stability, nuclear export and translation of mRNA transcripts. Control of poly(A) tail length is thus an important mechanism to regulate the abundance and ultimate translation of transcripts. Understanding the global regulation of poly(A) tail length will require dissecting the contributions of enzymes, regulatory factors, and poly(A) binding proteins (Pabs) that all cooperate to regulate polyadenylation. A recent addition to the Pab family is the CCCH-type zinc finger class of Pabs that includes S. cerevisiae Nab2 and its human counterpart, ZC3H14. In S. cerevisiae, Nab2 is an essential nuclear Pab implicated in both poly(A) RNA export from the nucleus and control of poly(A) tail length. Consistent with an important role in regulation of poly(A) tail length, depletion of Nab2 from yeast cells results in hyperadenylation of poly(A) RNA. In this review, we focus on the role of Nab2 in poly(A) tail length control and speculate on potential mechanisms by which Nab2 could regulate poly(A) tail length based on reported physical and genetic interactions. We present models, illustrating how Nab2 could regulate poly(A) tail length by limiting polyadenylation and/or enhancing trimming. Given that mutation of the gene encoding the human Nab2 homologue, ZC3H14, causes a form of autosomal recessive intellectual disability, we also speculate on how mutations in a gene encoding a ubiquitously expressed Pab lead specifically to neurological defects. This article is part of a Special Issue entitled: Nuclear Transport and RNA Processing.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Structure-function relationships in the Nab2 polyadenosine-RNA binding Zn finger protein family.Protein Sci. 2019 Mar;28(3):513-523. doi: 10.1002/pro.3565. Epub 2019 Jan 16. Protein Sci. 2019. PMID: 30578643 Free PMC article. Review.
-
Recognition of polyadenosine RNA by the zinc finger domain of nuclear poly(A) RNA-binding protein 2 (Nab2) is required for correct mRNA 3'-end formation.J Biol Chem. 2010 Aug 20;285(34):26022-32. doi: 10.1074/jbc.M110.141127. Epub 2010 Jun 16. J Biol Chem. 2010. PMID: 20554526 Free PMC article.
-
Structural basis for the dimerization of Nab2 generated by RNA binding provides insight into its contribution to both poly(A) tail length determination and transcript compaction in Saccharomyces cerevisiae.Nucleic Acids Res. 2017 Feb 17;45(3):1529-1538. doi: 10.1093/nar/gkw1224. Nucleic Acids Res. 2017. PMID: 28180315 Free PMC article.
-
Structural basis for polyadenosine-RNA binding by Nab2 Zn fingers and its function in mRNA nuclear export.Structure. 2012 Jun 6;20(6):1007-18. doi: 10.1016/j.str.2012.03.011. Epub 2012 May 3. Structure. 2012. PMID: 22560733 Free PMC article.
-
Birth of a poly(A) tail: mechanisms and control of mRNA polyadenylation.FEBS Open Bio. 2023 Jul;13(7):1140-1153. doi: 10.1002/2211-5463.13528. Epub 2022 Dec 7. FEBS Open Bio. 2023. PMID: 36416579 Free PMC article. Review.
Cited by
-
Determinants and implications of mRNA poly(A) tail size--does this protein make my tail look big?Semin Cell Dev Biol. 2014 Oct;34:24-32. doi: 10.1016/j.semcdb.2014.05.018. Epub 2014 Jun 5. Semin Cell Dev Biol. 2014. PMID: 24910447 Free PMC article. Review.
-
Unraveling docking and initiation of mRNA export through the nuclear pore complex.Bioessays. 2022 Aug;44(8):e2200027. doi: 10.1002/bies.202200027. Epub 2022 Jun 26. Bioessays. 2022. PMID: 35754154 Free PMC article.
-
The choreography of chromatin in RNA polymerase III regulation.Biochem Soc Trans. 2024 Jun 26;52(3):1173-1189. doi: 10.1042/BST20230770. Biochem Soc Trans. 2024. PMID: 38666598 Free PMC article. Review.
-
Polyadenylation and nuclear export of mRNAs.J Biol Chem. 2019 Mar 1;294(9):2977-2987. doi: 10.1074/jbc.REV118.005594. Epub 2019 Jan 25. J Biol Chem. 2019. PMID: 30683695 Free PMC article. Review.
-
The yeast Ty1 retrotransposon requires components of the nuclear pore complex for transcription and genomic integration.Nucleic Acids Res. 2018 Apr 20;46(7):3552-3578. doi: 10.1093/nar/gky109. Nucleic Acids Res. 2018. PMID: 29514267 Free PMC article.
References
-
- Moore MJ, Proudfoot NJ. Cell. 2009;136:688–700. - PubMed
-
- Proudfoot NJ, Furger A, Dye MJ. Cell. 2002;108:501–512. - PubMed
-
- Schmid M, Jensen TH. Wiley Interdiscip Rev RNA. 2010;1:474–485. - PubMed
-
- Doma MK, Parker R. Cell. 2007;131:660–668. - PubMed
-
- Lebreton A, Seraphin B. Biochim Biophys Acta. 2008;1779:558–565. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

