Nkx3.1 and Myc crossregulate shared target genes in mouse and human prostate tumorigenesis
- PMID: 22484818
- PMCID: PMC3336973
- DOI: 10.1172/JCI58540
Nkx3.1 and Myc crossregulate shared target genes in mouse and human prostate tumorigenesis
Abstract
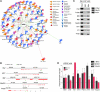
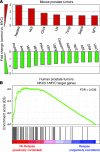
Cooperativity between oncogenic mutations is recognized as a fundamental feature of malignant transformation, and it may be mediated by synergistic regulation of the expression of pro- and antitumorigenic target genes. However, the mechanisms by which oncogenes and tumor suppressors coregulate downstream targets and pathways remain largely unknown. Here, we used ChIP coupled to massively parallel sequencing (ChIP-seq) and gene expression profiling in mouse prostates to identify direct targets of the tumor suppressor Nkx3.1. Further analysis indicated that a substantial fraction of Nkx3.1 target genes are also direct targets of the oncoprotein Myc. We also showed that Nkx3.1 and Myc bound to and crossregulated shared target genes in mouse and human prostate epithelial cells and that Nkx3.1 could oppose the transcriptional activity of Myc. Furthermore, loss of Nkx3.1 cooperated with concurrent overexpression of Myc to promote prostate cancer in transgenic mice. In human prostate cancer patients, dysregulation of shared NKX3.1/MYC target genes was associated with disease relapse. Our results indicate that NKX3.1 and MYC coregulate prostate tumorigenesis by converging on, and crossregulating, a common set of target genes. We propose that coregulation of target gene expression by oncogenic/tumor suppressor transcription factors may represent a general mechanism underlying the cooperativity of oncogenic mutations during tumorigenesis.
Figures







Comment in
-
Re: Nkx3.1 and myc crossregulate shared target genes in mouse and human prostate tumorigenesis.J Urol. 2013 Feb;189(2):772. doi: 10.1016/j.juro.2012.10.109. Epub 2012 Dec 20. J Urol. 2013. PMID: 23312205 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

