Skip the alignment: degenerate, multiplex primer and probe design using K-mer matching instead of alignments
- PMID: 22485178
- PMCID: PMC3317645
- DOI: 10.1371/journal.pone.0034560
Skip the alignment: degenerate, multiplex primer and probe design using K-mer matching instead of alignments
Abstract
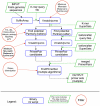
PriMux is a new software package for selecting multiplex compatible, degenerate primers and probes to detect diverse targets such as viruses. It requires no multiple sequence alignment, instead applying k-mer algorithms, hence it scales well for large target sets and saves user effort from curating sequences into alignable groups. PriMux has the capability to predict degenerate primers as well as probes suitable for TaqMan or other primer/probe triplet assay formats, or simply probes for microarray or other single-oligo assay formats. PriMux employs suffix array methods for efficient calculations on oligos 10-~100 nt in length. TaqMan® primers and probes for each segment of Rift Valley fever virus were designed using PriMux, and lab testing comparing signatures designed using PriMux versus those designed using traditional methods demonstrated equivalent or better sensitivity for the PriMux-designed signatures compared to traditional signatures. In addition, we used PriMux to design TaqMan® primers and probes for unalignable or poorly alignable groups of targets: that is, all segments of Rift Valley fever virus analyzed as a single target set of 198 sequences, or all 2863 Dengue virus genomes for all four serotypes available at the time of our analysis. The PriMux software is available as open source from http://sourceforge.net/projects/PriMux.
Conflict of interest statement
Figures
References
-
- Lemmon GH, Gardner SN. Predicting the sensitivity and specificity of published real-time PCR assays. Ann Clin Microbiol Antimicrob. 2008;7:18. doi: 10.1186/1476-0711-1187-1118. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous