A competitive nucleotide binding inhibitor: in vitro characterization of Rab7 GTPase inhibition
- PMID: 22486388
- PMCID: PMC3440014
- DOI: 10.1021/cb3001099
A competitive nucleotide binding inhibitor: in vitro characterization of Rab7 GTPase inhibition
Abstract
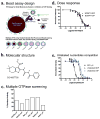
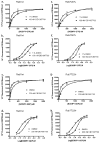
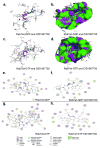
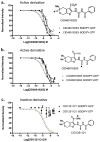
Mapping the functionality of GTPases through small molecule inhibitors represents an underexplored area in large part due to the lack of suitable compounds. Here we report on the small chemical molecule 2-(benzoylcarbamothioylamino)-5,5-dimethyl-4,7-dihydrothieno[2,3-c]pyran-3-carboxylic acid (PubChem CID 1067700) as an inhibitor of nucleotide binding by Ras-related GTPases. The mechanism of action of this pan-GTPase inhibitor was characterized in the context of the Rab7 GTPase as there are no known inhibitors of Rab GTPases. Bead-based flow cytometry established that CID 1067700 has significant inhibitory potency on Rab7 nucleotide binding with nanomolar inhibitor (K(i)) values and an inhibitory response of ≥97% for BODIPY-GTP and BODIPY-GDP binding. Other tested GTPases exhibited significantly lower responses. The compound behaves as a competitive inhibitor of Rab7 nucleotide binding based on both equilibrium binding and dissociation assays. Molecular docking analyses are compatible with CID 1067700 fitting into the nucleotide binding pocket of the GTP-conformer of Rab7. On the GDP-conformer, the molecule has greater solvent exposure and significantly less protein interaction relative to GDP, offering a molecular rationale for the experimental results. Structural features pertinent to CID 1067700 inhibitory activity have been identified through initial structure-activity analyses and identified a molecular scaffold that may serve in the generation of more selective probes for Rab7 and other GTPases. Taken together, our study has identified the first competitive GTPase inhibitor and demonstrated the potential utility of the compound for dissecting the enzymology of the Rab7 GTPase, as well as serving as a model for other small molecular weight GTPase inhibitors.
Figures

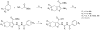
References
-
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54MH074425/MH/NIMH NIH HHS/United States
- U54HG005031/HG/NHGRI NIH HHS/United States
- U54 MH084690/MH/NIMH NIH HHS/United States
- UL1 TR001449/TR/NCATS NIH HHS/United States
- P30CA1181000/CA/NCI NIH HHS/United States
- R21NS066435/NS/NINDS NIH HHS/United States
- U54 HG005031/HG/NHGRI NIH HHS/United States
- R21 NS066435/NS/NINDS NIH HHS/United States
- R03MH081231/MH/NIMH NIH HHS/United States
- U54 MH074425/MH/NIMH NIH HHS/United States
- UL1 TR000041/TR/NCATS NIH HHS/United States
- U54MH084690/MH/NIMH NIH HHS/United States
- R03 MH081231/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

