RNA plasticity and selectivity applicable to therapeutics and novel biosensor development
- PMID: 22487172
- PMCID: PMC3444689
- DOI: 10.1111/j.1365-2443.2012.01596.x
RNA plasticity and selectivity applicable to therapeutics and novel biosensor development
Abstract
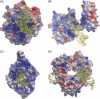
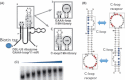
Aptamers are short, single-stranded nucleic acid sequences that are selected in vitro from large oligonucleotide libraries based on their high affinity to a target molecule. Hence, aptamers can be thought of as a nucleic acid analog to antibodies. However, several viewpoints hold that the potential of aptamers arises from interesting characteristics that are distinct from, or in some cases, superior to those of antibodies. This review summarizes the recent achievements in aptamer programs developed in our laboratory against basic and therapeutic protein targets. Through these studies, we became aware of the remarkable conformational plasticity and selectivity of RNA, on which the published report has not shed much light even though this is evidently a crucial feature for the strong specificity and affinity of RNA aptamers.
© 2012 The Authors. Journal compilation © 2012 by the Molecular Biology Society of Japan/Blackwell Publishing Ltd.
Figures




References
-
- Adachi H, Ishiguro A, Hamada M, Sakota E, Asai K, Nakamura Y. Antagonistic RNA aptamer specific to a heterodimeric form of human interleukin-17A/F. Biochimie. 2011;93:1081–1088. - PubMed
-
- Akiyama H, Kachi S, Silva RL, Umeda N, Hackett SF, McCauley D, McCauley T, Zoltoski A, Epstein DM, Campochiaro PA. Intraocular injection of an aptamer that binds PDGF-B: a potential treatment for proliferative retinopathies. J. Cell. Physiol. 2006;207:407–412. - PubMed
-
- Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes. J. Am. Chem. Soc. 2003;125:14716–14717. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

