Tropomodulins: pointed-end capping proteins that regulate actin filament architecture in diverse cell types
- PMID: 22488942
- PMCID: PMC3444156
- DOI: 10.1002/cm.21031
Tropomodulins: pointed-end capping proteins that regulate actin filament architecture in diverse cell types
Abstract
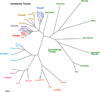
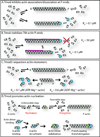
Tropomodulins are a family of four proteins (Tmods 1-4) that cap the pointed ends of actin filaments in actin cytoskeletal structures in a developmentally regulated and tissue-specific manner. Unique among capping proteins, Tmods also bind tropomyosins (TMs), which greatly enhance the actin filament pointed-end capping activity of Tmods. Tmods are defined by a TM-regulated/Pointed-End Actin Capping (TM-Cap) domain in their unstructured N-terminal portion, followed by a compact, folded Leucine-Rich Repeat/Pointed-End Actin Capping (LRR-Cap) domain. By inhibiting actin monomer association and dissociation from pointed ends, Tmods regulate actin dynamics and turnover, stabilizing actin filament lengths and cytoskeletal architecture. In this review, we summarize the genes, structural features, molecular and biochemical properties, actin regulatory mechanisms, expression patterns, and cell and tissue functions of Tmods. By understanding Tmods' functions in the context of their molecular structure, actin regulation, binding partners, and related variants (leiomodins 1-3), we can draw broad conclusions that can explain the diverse morphological and functional phenotypes that arise from Tmod perturbation experiments in vitro and in vivo. Tmod-based stabilization and organization of intracellular actin filament networks provide key insights into how the emergent properties of the actin cytoskeleton drive tissue morphogenesis and physiology.
Copyright © 2012 Wiley Periodicals, Inc.
Figures




References
-
- Alizadeh A, Clark J, Seeberger T, Hess J, Blankenship T, FitzGerald PG. Targeted deletion of the lens fiber cell-specific intermediate filament protein filensin. Invest Ophthalmol Vis Sci. 2003;44(12):5252–5258. - PubMed
-
- Allen DP, Low PS, Dola A, Maisel H. Band 3 and ankyrin homologues are present in eye lens: evidence for all major erythrocyte membrane components in same non-erythroid cell. Biochem Biophys Res Commun. 1987;149(1):266–275. - PubMed
-
- Almenar-Queralt A, Gregorio CC, Fowler VM. Tropomodulin assembles early in myofibrillogenesis in chick skeletal muscle: evidence that thin filaments rearrange to form striated myofibrils. J Cell Sci. 1999a;112(Pt 8):1111–1123. - PubMed
-
- Almenar-Queralt A, Lee A, Conley CA, Ribas de Pouplana L, Fowler VM. Identification of a novel tropomodulin isoform, skeletal tropomodulin, that caps actin filament pointed ends in fast skeletal muscle. J Biol Chem. 1999b;274(40):28466–28475. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

