Radiation-induced reprogramming of breast cancer cells
- PMID: 22489015
- PMCID: PMC3413333
- DOI: 10.1002/stem.1058
Radiation-induced reprogramming of breast cancer cells
Abstract
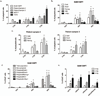
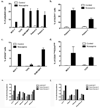
Breast cancers are thought to be organized hierarchically with a small number of breast cancer stem cells (BCSCs) able to regrow a tumor while their progeny lack this ability. Recently, several groups reported enrichment for BCSCs when breast cancers were subjected to classic anticancer treatment. However, the underlying mechanisms leading to this enrichment are incompletely understood. Using non-BCSCs sorted from patient samples, we found that ionizing radiation reprogrammed differentiated breast cancer cells into induced BCSCs (iBCSCs). iBCSCs showed increased mammosphere formation, increased tumorigenicity, and expressed the same stemness-related genes as BCSCs from nonirradiated samples. Reprogramming occurred in a polyploid subpopulation of cells, coincided with re-expression of the transcription factors Oct4, sex determining region Y-box 2, Nanog, and Klf4, and could be partially prevented by Notch inhibition. We conclude that radiation may induce a BCSC phenotype in differentiated breast cancer cells and that this mechanism contributes to increased BCSC numbers seen after classic anticancer treatment.
Copyright © 2012 AlphaMed Press.
Figures





References
-
- Marcato P, Dean CA, Pan D, et al. Aldehyde Dehydrogenase Activity of Breast Cancer Stem Cells is Primarily Due to Isoform ALDH1A3 and Its Expression is Predictive of Metastasis. Stem Cells. 2010 - PubMed
-
- Phillips TM, McBride WH, Pajonk F. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98:1777–1785. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

