Scanometric microRNA array profiling of prostate cancer markers using spherical nucleic acid-gold nanoparticle conjugates
- PMID: 22489825
- PMCID: PMC3357313
- DOI: 10.1021/ac3004055
Scanometric microRNA array profiling of prostate cancer markers using spherical nucleic acid-gold nanoparticle conjugates
Abstract
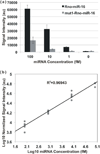
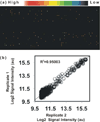
We report the development of a novel Scanometric MicroRNA (Scano-miR) platform for the detection of relatively low abundance miRNAs with high specificity and reproducibility. The Scano-miR system was able to detect 1 fM concentrations of miRNA in serum with single nucleotide mismatch specificity. Indeed, it provides increased sensitivity for miRNA targets compared to molecular fluorophore-based detection systems, where 88% of the low abundance miRNA targets could not be detected under identical conditions. The application of the Scano-miR platform to high density array formats demonstrates its utility for high throughput and multiplexed miRNA profiling from various biological samples. To assess the accuracy of the Scano-miR system, we analyzed the miRNA profiles of samples from men with prostate cancer (CaP), the most common noncutaneous malignancy and the second leading cause of cancer death among American men. The platform exhibits 98.8% accuracy when detecting deregulated miRNAs involved in CaP, which demonstrates its potential utility in profiling and identifying clinical and research biomarkers.
Figures

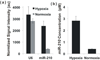

References
-
- Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. Nat. Rev. Cancer. 2009;9:293. - PubMed
-
- Lee RC, Feinbaum RL, Ambros V. Cell. 1993;75:843. - PubMed
-
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Nature. 2005;433:769. - PubMed
-
- Lewis BP, Burge CB, Bartel DP. Cell. 2005;120:15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

