Tracking dynamics of plant biomass composting by changes in substrate structure, microbial community, and enzyme activity
- PMID: 22490508
- PMCID: PMC3384452
- DOI: 10.1186/1754-6834-5-20
Tracking dynamics of plant biomass composting by changes in substrate structure, microbial community, and enzyme activity
Abstract
Background: Understanding the dynamics of the microbial communities that, along with their secreted enzymes, are involved in the natural process of biomass composting may hold the key to breaking the major bottleneck in biomass-to-biofuels conversion technology, which is the still-costly deconstruction of polymeric biomass carbohydrates to fermentable sugars.However, the complexity of both the structure of plant biomass and its counterpart microbial degradation communities makes it difficult to investigate the composting process.
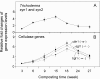
Results: In this study, a composter was set up with a mix of yellow poplar (Liriodendron tulipifera) wood-chips and mown lawn grass clippings (85:15 in dry-weight) and used as a model system. The microbial rDNA abundance data obtained from analyzing weekly-withdrawn composted samples suggested population-shifts from bacteria-dominated to fungus-dominated communities. Further analyses by an array of optical microscopic, transcriptional and enzyme-activity techniques yielded correlated results, suggesting that such population shifts occurred along with early removal of hemicellulose followed by attack on the consequently uncovered cellulose as the composting progressed.
Conclusion: The observed shifts in dominance by representative microbial groups, along with the observed different patterns in the gene expression and enzymatic activities between cellulases, hemicellulases, and ligninases during the composting process, provide new perspectives for biomass-derived biotechnology such as consolidated bioprocessing (CBP) and solid-state fermentation for the production of cellulolytic enzymes and biofuels.
Figures





References
-
- Ryckeboer J, Mergaert J, Vaes K, Klammer S, De Clercq D, Coosemans J, Insam H, Swings J. A survey of bacteria and fungi occurring during composting and self-heating processes. Annals of Microbiology. 2003;53:349–410.
-
- Wei H, Luo Y, Guo J, Yan L, Yang B, Zhang J, Hu Z, Sun L. Technological studies on production of biological-organic-inorganic compound fertilizer by utilizing the manure of livestock and poultry. Biotechnology. 1999;9:30–34.
-
- Wei H, Guo J, Zhou Q, Wang Z, Wu Z, Zhang J. Studies on biological-organic-inorganic fertilizer and its field application. Journal of Microbiology. 1997;17:18–24.
LinkOut - more resources
Full Text Sources

