Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides
- PMID: 22491177
- PMCID: PMC3422608
- DOI: 10.1038/mi.2012.23
Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides
Abstract
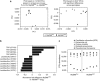
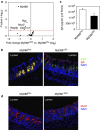
Intestinal epithelial cells (IECs) form a physical and immunological barrier that separates the vast gut microbiota from host tissues. MyD88-dependent Toll-like receptor signaling is a key mediator of microbial-host cross-talk. We examined the role of epithelial MyD88 expression by generating mice with an IEC-targeted deletion of the Myd88 gene (MyD88(ΔIEC)). Loss of epithelial MyD88 signaling resulted in increased numbers of mucus-associated bacteria; translocation of bacteria, including the opportunistic pathogen Klebsiella pneumoniae, to mesenteric lymph nodes; reduced transmucosal electrical resistance; impaired mucus-associated antimicrobial activity; and downregulated expression of polymeric immunoglobulin receptor (the epithelial IgA transporter), mucin-2 (the major protein of intestinal mucus), and the antimicrobial peptides RegIIIγ and Defa-rs1. We further observed significant differences in the composition of the gut microbiota between MyD88(ΔIEC) mice and wild-type littermates. These physical, immunological, and microbial defects resulted in increased susceptibility of MyD88(ΔIEC) mice to experimental colitis. We conclude that MyD88 signaling in IECs is crucial for maintenance of gut homeostasis.
Figures




References
-
- Hooper L.V., Macpherson A.J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010;10,:159–169. - PubMed
-
- Carvalho F.A., Aitken J.D., Vijay-Kumar M., Gewirtz A.T. Toll-like receptor-gut microbiota interactions: perturb at your own risk! Annu. Rev. Physiol. 2011;74,:177–198. - PubMed
-
- Gill N., Wlodarska M., Finlay B.B. Roadblocks in the gut: barriers to enteric infection. Cell. Microbiol. 2011;13,:660–669. - PubMed
-
- Cario E. Heads up! How the intestinal epithelium safeguards mucosal barrier immunity through the inflammasome and beyond. Curr. Opin. Gastroenterol. 2010;26,:583–590. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

