Population genetics models of local ancestry
- PMID: 22491189
- PMCID: PMC3374321
- DOI: 10.1534/genetics.112.139808
Population genetics models of local ancestry
Abstract
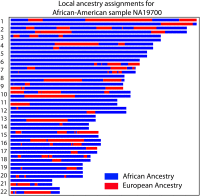
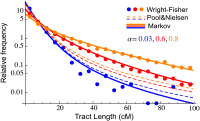
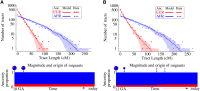
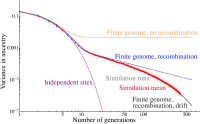
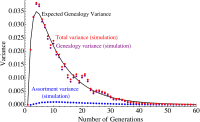
Migrations have played an important role in shaping the genetic diversity of human populations. Understanding genomic data thus requires careful modeling of historical gene flow. Here we consider the effect of relatively recent population structure and gene flow and interpret genomes of individuals that have ancestry from multiple source populations as mosaics of segments originating from each population. This article describes general and tractable models for local ancestry patterns with a focus on the length distribution of continuous ancestry tracts and the variance in total ancestry proportions among individuals. The models offer improved agreement with Wright-Fisher simulation data when compared to the state-of-the art and can be used to infer time-dependent migration rates from multiple populations. Considering HapMap African-American (ASW) data, we find that a model with two distinct phases of "European" gene flow significantly improves the modeling of both tract lengths and ancestry variances.
Figures

References
-
- Bercovici S., Geiger D., 2009. Inferring ancestries efficiently in admixed populations with linkage disequilibrium. J. Comput. Biol. 16(8): 1141–1150 - PubMed
-
- Brisbin, A., 2010 Linkage analysis for categorical traits and ancestry assignment in admixed individuals. Ph.D. Thesis, Cornell University, Ithaca, NY.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

