The removal of RNA primers from DNA synthesized by the reverse transcriptase of the retrotransposon Tf1 is stimulated by Tf1 integrase
- PMID: 22491446
- PMCID: PMC3372178
- DOI: 10.1128/JVI.00009-12
The removal of RNA primers from DNA synthesized by the reverse transcriptase of the retrotransposon Tf1 is stimulated by Tf1 integrase
Abstract
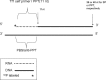
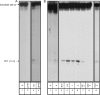
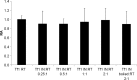
The Tf1 retrotransposon represents a group of long terminal repeat retroelements that use an RNA self-primer for initiating reverse transcription while synthesizing the minus-sense DNA strand. Tf1 reverse transcriptase (RT) was found earlier to generate the self-primer in vitro. Here, we show that this RT can remove from the synthesized cDNA the entire self-primer as well as the complete polypurine tract (PPT) sequence (serving as a second primer for cDNA synthesis). However, these primer removals, mediated by the RNase H activity of Tf1 RT, are quite inefficient. Interestingly, the integrase of Tf1 stimulated the specific Tf1 RT-directed cleavage of both the self-primer and PPT, although there was no general enhancement of the RT's RNase H activity (and the integrase by itself is devoid of any primer cleavage). The RTs of two prototype retroviruses, murine leukemia virus and human immunodeficiency virus, showed only a partial and nonspecific cleavage of both Tf1-associated primers with no stimulation by Tf1 integrase. Mutagenesis of Tf1 integrase revealed that the complete Tf1 integrase protein (excluding its chromodomain) is required for stimulating the Tf1 RT primer removal activity. Nonetheless, a double mutant integrase that has lost its integration functions can still stimulate the RT's activity, though heat-inactivated integrase cannot enhance primer removals. These findings suggest that the enzymatic activity of Tf1 integrase is not essential for stimulating the RT-mediated primer removal, while the proper folding of this protein is obligatory for this function. These results highlight possible new functions of Tf1 integrase in the retrotransposon's reverse transcription process.
Figures





References
-
- Avidan O, Loya S, Tonjes RR, Sevilya Z, Hizi A. 2003. Expression and characterization of a recombinant novel reverse transcriptase of a porcine endogenous retrovirus. Virology 307:341–357 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

