Serology-enabled discovery of genetically diverse hepaciviruses in a new host
- PMID: 22491452
- PMCID: PMC3372197
- DOI: 10.1128/JVI.00250-12
Serology-enabled discovery of genetically diverse hepaciviruses in a new host
Abstract
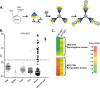
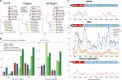
Genetic and biological characterization of new hepaciviruses infecting animals contributes to our understanding of the ultimate origins of hepatitis C virus (HCV) infection in humans and dramatically enhances our ability to study its pathogenesis using tractable animal models. Animal homologs of HCV include a recently discovered canine hepacivirus (CHV) and GB virus B (GBV-B), both viruses with largely undetermined natural host ranges. Here we used a versatile serology-based approach to determine the natural host of the only known nonprimate hepacivirus (NPHV), CHV, which is also the closest phylogenetic relative of HCV. Recombinant protein expressed from the helicase domain of CHV NS3 was used as antigen in the luciferase immunoprecipitation system (LIPS) assay to screen several nonprimate animal species. Thirty-six samples from 103 horses were immunoreactive, and viral genomic RNA was present in 8 of the 36 seropositive animals and none of the seronegative animals. Complete genome sequences of these 8 genetically diverse NPHVs showed 14% (range, 6.4% to 17.2%) nucleotide sequence divergence, with most changes occurring at synonymous sites. RNA secondary structure prediction of the 383-base 5' untranslated region of NPHV was refined and extended through mapping of polymorphic sites to unpaired regions or (semi)covariant pairings. Similar approaches were adopted to delineate extensive RNA secondary structures in the coding region of the genome, predicted to form 27 regularly spaced, thermodynamically stable stem-loops. Together, these findings suggest a promising new nonprimate animal model and provide a database that will aid creation of functional NPHV cDNA clones and other novel tools for hepacivirus studies.
Figures

References
-
- Bruderer U, et al. 2004. Differentiating infection from vaccination in foot-and-mouth-disease: evaluation of an ELISA based on recombinant 3ABC. Vet. Microbiol. 101:187–197 - PubMed
-
- Bukh J, Apgar CL, Govindarajan S, Purcell RH. 2001. Host range studies of GB virus-B hepatitis agent, the closest relative of hepatitis C virus, in New World monkeys and chimpanzees. J. Med. Virol. 65:694–697 - PubMed
-
- Bukh J, Apgar CL, Yanagi M. 1999. Toward a surrogate model for hepatitis C virus: an infectious molecular clone of the GB virus-B hepatitis agent. Virology 262:470–478 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- R21 AI081132/AI/NIAID NIH HHS/United States
- AI070411/AI/NIAID NIH HHS/United States
- U01 AI070411/AI/NIAID NIH HHS/United States
- R24 EY017404/EY/NEI NIH HHS/United States
- 095831/Wellcome Trust/United Kingdom
- U54 AI057158/AI/NIAID NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- AI081132/AI/NIAID NIH HHS/United States
- EY017404/EY/NEI NIH HHS/United States
- R21 AI090196/AI/NIAID NIH HHS/United States
- R01 AI079231/AI/NIAID NIH HHS/United States
- AI57158/AI/NIAID NIH HHS/United States
- AI079231/AI/NIAID NIH HHS/United States
- AI090196/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

