Combining H/D exchange mass spectroscopy and computational docking reveals extended DNA-binding surface on uracil-DNA glycosylase
- PMID: 22492624
- PMCID: PMC3401472
- DOI: 10.1093/nar/gks291
Combining H/D exchange mass spectroscopy and computational docking reveals extended DNA-binding surface on uracil-DNA glycosylase
Abstract
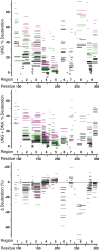
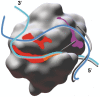
X-ray crystallography provides excellent structural data on protein-DNA interfaces, but crystallographic complexes typically contain only small fragments of large DNA molecules. We present a new approach that can use longer DNA substrates and reveal new protein-DNA interactions even in extensively studied systems. Our approach combines rigid-body computational docking with hydrogen/deuterium exchange mass spectrometry (DXMS). DXMS identifies solvent-exposed protein surfaces; docking is used to create a 3-dimensional model of the protein-DNA interaction. We investigated the enzyme uracil-DNA glycosylase (UNG), which detects and cleaves uracil from DNA. UNG was incubated with a 30 bp DNA fragment containing a single uracil, giving the complex with the abasic DNA product. Compared with free UNG, the UNG-DNA complex showed increased solvent protection at the UNG active site and at two regions outside the active site: residues 210-220 and 251-264. Computational docking also identified these two DNA-binding surfaces, but neither shows DNA contact in UNG-DNA crystallographic structures. Our results can be explained by separation of the two DNA strands on one side of the active site. These non-sequence-specific DNA-binding surfaces may aid local uracil search, contribute to binding the abasic DNA product and help present the DNA product to APE-1, the next enzyme on the DNA-repair pathway.
Figures
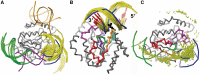
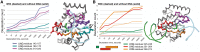
References
-
- Tan S, Hunziker Y, Pellegrini L, Richmond TJ. Crystallization of the yeast MATα2/MCM1/DNA ternary complex: general methods and principles for protein/DNA cocrystallization. J. Mol. Biol. 2000;297:947–959. - PubMed
-
- Janin J, Henrick K, Moult J, Ten Eyck L, Sternberg MJE, Vajda S, Vakser I, Wodak SJ. CAPRI: A Critical Assessment of PRedicted Interactions. Proteins. 2003;52:2–9. - PubMed
-
- Méndez R, Leplae R, De Maria L, Wodak SJ. Assessment of blind predictions of protein-protein interactions: Current status of docking methods. Proteins. 2003;52:51–67. - PubMed
-
- Méndez R, Leplae R, Lensink MF, Wodak SJ. Assessment of CAPRI predictions in rounds 3-5 shows progress in docking procedures. Proteins. 2005;60:150–169. - PubMed
-
- Giudice E, Lavery R. Simulations of nucleic acids and their complexes. Acc. Chem. Res. 2002;35:350–357. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM037684/GM/NIGMS NIH HHS/United States
- AI081982/AI/NIAID NIH HHS/United States
- CA099835/CA/NCI NIH HHS/United States
- AI068730/AI/NIAID NIH HHS/United States
- R01 GM020501/GM/NIGMS NIH HHS/United States
- AI072106/AI/NIAID NIH HHS/United States
- GM020501/GM/NIGMS NIH HHS/United States
- GM066170/GM/NIGMS NIH HHS/United States
- AI2008031/AI/NIAID NIH HHS/United States
- R01 GM046312/GM/NIGMS NIH HHS/United States
- AI076961/AI/NIAID NIH HHS/United States
- CA118595/CA/NCI NIH HHS/United States
- GM070996/GM/NIGMS NIH HHS/United States
- GM46312/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

