SAVoR: a server for sequencing annotation and visualization of RNA structures
- PMID: 22492627
- PMCID: PMC3394343
- DOI: 10.1093/nar/gks310
SAVoR: a server for sequencing annotation and visualization of RNA structures
Abstract
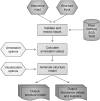
RNA secondary structure is required for the proper regulation of the cellular transcriptome. This is because the functionality, processing, localization and stability of RNAs are all dependent on the folding of these molecules into intricate structures through specific base pairing interactions encoded in their primary nucleotide sequences. Thus, as the number of RNA sequencing (RNA-seq) data sets and the variety of protocols for this technology grow rapidly, it is becoming increasingly pertinent to develop tools that can analyze and visualize this sequence data in the context of RNA secondary structure. Here, we present Sequencing Annotation and Visualization of RNA structures (SAVoR), a web server, which seamlessly links RNA structure predictions with sequencing data and genomic annotations to produce highly informative and annotated models of RNA secondary structure. SAVoR accepts read alignment data from RNA-seq experiments and computes a series of per-base values such as read abundance and sequence variant frequency. These values can then be visualized on a customizable secondary structure model. SAVoR is freely available at http://tesla.pcbi.upenn.edu/savor.
Figures


References
-
- Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. - PubMed
-
- Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. - PubMed
-
- Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell. 2011;43:340–352. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

