An analysis of substitution, deletion and insertion mutations in cancer genes
- PMID: 22492711
- PMCID: PMC3413105
- DOI: 10.1093/nar/gks290
An analysis of substitution, deletion and insertion mutations in cancer genes
Abstract
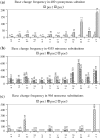
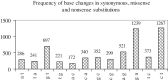
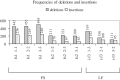
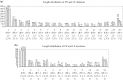
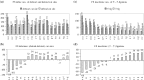
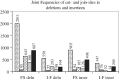
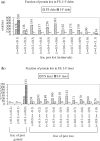
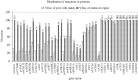
Cancer-associated mutations in cancer genes constitute a diverse set of mutations associated with the disease. To gain insight into features of the set, substitution, deletion and insertion mutations were analysed at the nucleotide level, from the COSMIC database. The most frequent substitutions were c → t, g → a, g → t, and the most frequent codon changes were to termination codons. Deletions more than insertions, FS (frameshift) indels more than I-F (in-frame) ones, and single-nucleotide indels, were frequent. FS indels cause loss of significant fractions of proteins. The 5'-cut in FS deletions, and 5'-ligation in FS insertions, often occur between pairs of identical bases. Interestingly, the cut-site and 3'-ligation in insertions, and 3'-cut and join-pair in deletions, were each found to be the same significantly often (p < 0.001). It is suggested that these features aid the incorporation of indel mutations. Tumor suppressors undergo larger numbers of mutations, especially disruptive ones, over the entire protein length, to inactivate two alleles. Proto-oncogenes undergo fewer, less-disruptive mutations, in selected protein regions, to activate a single allele. Finally, catalogues, in ranked order, of genes mutated in each cancer, and cancers in which each gene is mutated, were created. The study highlights the nucleotide level preferences and disruptive nature of cancer mutations.
Figures
References
-
- Stehelin D, Varmus HE, Bishop JM, Vogt PK. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature. 1976;260:170–173. - PubMed
-
- Tabin CJ, Bradley SM, Bargmann CI, Weinberg RA, Papageorge AG, Scolnick EM, Dhar R, Lowy DR, Chang EH. Mechanism of activation of a human oncogene. Nature. 1982;300:143–9. - PubMed
-
- Harris H, Miller OJ, Klein G, Worst P, Tachibana T. Suppression of malignancy by cell fusion. Nature. 1969;223:363–368. - PubMed
-
- Weinberg RA. Tumor suppressor genes. Science. 1991;254:1138–46. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

