Fructose degradation in the haloarchaeon Haloferax volcanii involves a bacterial type phosphoenolpyruvate-dependent phosphotransferase system, fructose-1-phosphate kinase, and class II fructose-1,6-bisphosphate aldolase
- PMID: 22493022
- PMCID: PMC3370872
- DOI: 10.1128/JB.00200-12
Fructose degradation in the haloarchaeon Haloferax volcanii involves a bacterial type phosphoenolpyruvate-dependent phosphotransferase system, fructose-1-phosphate kinase, and class II fructose-1,6-bisphosphate aldolase
Abstract
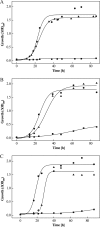
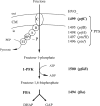
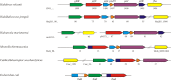
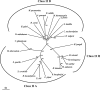
The halophilic archaeon Haloferax volcanii utilizes fructose as a sole carbon and energy source. Genes and enzymes involved in fructose uptake and degradation were identified by transcriptional analyses, deletion mutant experiments, and enzyme characterization. During growth on fructose, the gene cluster HVO_1495 to HVO_1499, encoding homologs of the five bacterial phosphotransferase system (PTS) components enzyme IIB (EIIB), enzyme I (EI), histidine protein (HPr), EIIA, and EIIC, was highly upregulated as a cotranscript. The in-frame deletion of HVO_1499, designated ptfC (ptf stands for phosphotransferase system for fructose) and encoding the putative fructose-specific membrane component EIIC, resulted in a loss of growth on fructose, which could be recovered by complementation in trans. Transcripts of HVO_1500 (pfkB) and HVO_1494 (fba), encoding putative fructose-1-phosphate kinase (1-PFK) and fructose-1,6-bisphosphate aldolase (FBA), respectively, as well as 1-PFK and FBA activities were specifically upregulated in fructose-grown cells. pfkB and fba knockout mutants did not grow on fructose, whereas growth on glucose was not inhibited, indicating the functional involvement of both enzymes in fructose catabolism. Recombinant 1-PFK and FBA obtained after homologous overexpression were characterized as having kinetic properties indicative of functional 1-PFK and a class II type FBA. From these data, we conclude that fructose uptake in H. volcanii involves a fructose-specific PTS generating fructose-1-phosphate, which is further converted via fructose-1,6-bisphosphate to triose phosphates by 1-PFK and FBA. This is the first report of the functional involvement of a bacterial-like PTS and of class II FBA in the sugar metabolism of archaea.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

