Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27
- PMID: 22493065
- PMCID: PMC3372260
- DOI: 10.1128/MCB.06392-11
Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27
Abstract
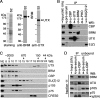
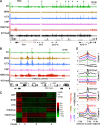
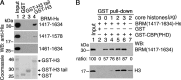
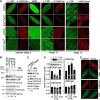
Trithorax group (TrxG) proteins antagonize Polycomb silencing and are required for maintenance of transcriptionally active states. We previously showed that the Drosophila melanogaster acetyltransferase CREB-binding protein (CBP) acetylates histone H3 lysine 27 (H3K27ac), thereby directly blocking its trimethylation (H3K27me3) by Polycomb repressive complex 2 (PRC2) in Polycomb target genes. Here, we show that H3K27ac levels also depend on other TrxG proteins, including the histone H3K27-specific demethylase UTX and the chromatin-remodeling ATPase Brahma (BRM). We show that UTX and BRM are physically associated with CBP in vivo and that UTX, BRM, and CBP colocalize genome-wide on Polycomb response elements (PREs) and on many active Polycomb target genes marked by H3K27ac. UTX and BRM bind directly to conserved zinc fingers of CBP, suggesting that their individual activities are functionally coupled in vivo. The bromodomain-containing C terminus of BRM binds to the CBP PHD finger, enhances PHD binding to histone H3, and enhances in vitro acetylation of H3K27 by recombinant CBP. brm mutations and knockdown of UTX by RNA interference (RNAi) reduce H3K27ac levels and increase H3K27me3 levels. We propose that direct binding of UTX and BRM to CBP and their modulation of H3K27ac play an important role in antagonizing Polycomb silencing.
Figures



References
-
- Agalioti T, et al. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103: 667– 678 - PubMed
-
- Agger K, et al. 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731– 734 - PubMed
-
- An W, Roeder RG. 2003. Direct association of p300 with unmodified H3 and H4 N termini modulates p300-dependent acetylation and transcription of nucleosomal templates. J. Biol. Chem. 278: 1504– 1510 - PubMed
-
- Bannister A, Kouzarides T. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384: 641– 643 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM039255/GM/NIGMS NIH HHS/United States
- S10 RR021228/RR/NCRR NIH HHS/United States
- S10 RR024536/RR/NCRR NIH HHS/United States
- R01HD056369/HD/NICHD NIH HHS/United States
- R01GM084947/GM/NIGMS NIH HHS/United States
- R01 HD056369/HD/NICHD NIH HHS/United States
- RR-021228-01/RR/NCRR NIH HHS/United States
- R01HG004722/HG/NHGRI NIH HHS/United States
- RR-024536-01/RR/NCRR NIH HHS/United States
- RR-017980-01/RR/NCRR NIH HHS/United States
- S10 RR017980/RR/NCRR NIH HHS/United States
- R01 HG004722/HG/NHGRI NIH HHS/United States
- R01GM39255/GM/NIGMS NIH HHS/United States
- R01 GM084947/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
