Drosophila Golgi membrane protein Ema promotes autophagosomal growth and function
- PMID: 22493244
- PMCID: PMC3344964
- DOI: 10.1073/pnas.1120320109
Drosophila Golgi membrane protein Ema promotes autophagosomal growth and function
Abstract
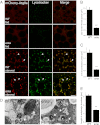
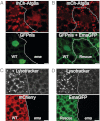
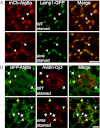
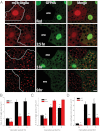
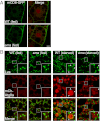
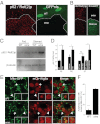
Autophagy is a self-degradative process in which cellular material is enclosed within autophagosomes and trafficked to lysosomes for degradation. Autophagosomal biogenesis is well described; however mechanisms controlling the growth and ultimate size of autophagosomes are unclear. Here we demonstrate that the Drosophila membrane protein Ema is required for the growth of autophagosomes. In an ema mutant, autophagosomes form in response to starvation and developmental cues, and these autophagosomes can mature into autolysosomes; however the autophagosomes are very small, and autophagy is impaired. In fat body cells, Ema localizes to the Golgi complex and is recruited to the membrane of autophagosomes in response to starvation. The Drosophila Golgi protein Lva also is recruited to the periphery of autophagosomes in response to starvation, and this recruitment requires ema. Therefore, we propose that Golgi is a membrane source for autophagosomal growth and that Ema facilitates this process. Clec16A, the human ortholog of Ema, is a candidate autoimmune susceptibility locus. Expression of Clec16A can rescue the autophagosome size defect in the ema mutant, suggesting that regulation of autophagosome morphogenesis may be a fundamental function of this gene family.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Suzuki K, Ohsumi Y. Current knowledge of the pre-autophagosomal structure (PAS) FEBS Lett. 2010;584:1280–1286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

