Functional asymmetries of proteasome translocase pore
- PMID: 22493437
- PMCID: PMC3365715
- DOI: 10.1074/jbc.M112.357327
Functional asymmetries of proteasome translocase pore
Abstract
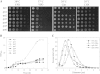
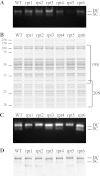
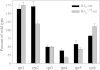
Degradation by proteasomes involves coupled translocation and unfolding of its protein substrates. Six distinct but paralogous proteasome ATPase proteins, Rpt1 to -6, form a heterohexameric ring that acts on substrates. An axially positioned loop (Ar-Φ loop) moves in concert with ATP hydrolysis, engages substrate, and propels it into a proteolytic chamber. The aromatic (Ar) residue of the Ar-Φ loop in all six Rpts of S. cerevisiae is tyrosine; this amino acid is thought to have important functional contacts with substrate. Six yeast strains were constructed and characterized in which Tyr was individually mutated to Ala. The mutant cells were viable and had distinct phenotypes. rpt3, rpt4, and rpt5 Tyr/Ala mutants, which cluster on one side of the ATPase hexamer, were substantially impaired in their capacity to degrade substrates. In contrast, rpt1, rpt2, and rpt6 mutants equaled or exceeded wild type in degradation activity. However, rpt1 and rpt6 mutants had defects that limited cell growth or viability under conditions that stressed the ubiquitin proteasome system. In contrast, the rpt3 mutant grew faster than wild type and to a smaller size, a defect that has previously been associated with misregulation of G1 cyclins. This rpt3 phenotype probably results from altered degradation of cell cycle regulatory proteins. Finally, mutation of five of the Rpt subunits increased proteasome ATPase activity, implying bidirectional coupling between the Ar-Φ loop and the ATP hydrolysis site. The present observations assign specific functions to individual Rpt proteins and provide insights into the diverse roles of the axial loops of individual proteasome ATPases.
Figures


References
-
- Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. (1997) Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 386, 463–471 - PubMed
-
- Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V. A., Finley D. (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94, 615–623 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

