Purification and identification of naringenin 7-O-methyltransferase, a key enzyme in biosynthesis of flavonoid phytoalexin sakuranetin in rice
- PMID: 22493492
- PMCID: PMC3365963
- DOI: 10.1074/jbc.M112.351270
Purification and identification of naringenin 7-O-methyltransferase, a key enzyme in biosynthesis of flavonoid phytoalexin sakuranetin in rice
Abstract
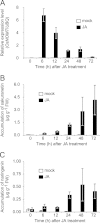
Sakuranetin, the major flavonoid phytoalexin in rice, is induced by ultraviolet (UV) irradiation, CuCl(2) treatment, jasmonic acid treatment, and infection by phytopathogens. It was recently demonstrated that sakuranetin has anti-inflammatory activity, anti-mutagenic activity, anti-pathogenic activities against Helicobacter pylori, Leishmania, and Trypanosoma and contributes to the maintenance of glucose homeostasis in animals. Thus, sakuranetin is a useful compound as a plant antibiotic and a potential pharmaceutical agent. Sakuranetin is biosynthesized from naringenin by naringenin 7-O-methyltransferase (NOMT). In previous research, rice NOMT (OsNOMT) was purified to apparent homogeneity from UV-treated wild-type rice leaves, but the purified protein, named OsCOMT1, exhibited caffeic acid O-methyltransferase (COMT) activity and not NOMT activity. In this study, we found that OsCOMT1 does not contribute to sakuranetin production in rice in vivo, and we purified OsNOMT using the oscomt1 mutant. A crude protein preparation from UV-treated oscomt1 leaves was subjected to three sequential purification steps, resulting in a 400-fold purification from the crude enzyme preparation. Using SDS-PAGE, the purest enzyme preparation showed a minor band at an apparent molecular mass of 40 kDa. Two O-methyltransferase-like proteins, encoded by Os04g0175900 and Os12g0240900, were identified from the 40-kDa band by MALDI-TOF/TOF analysis. Recombinant Os12g0240900 protein showed NOMT activity, but the recombinant Os04g0175900 protein did not. Os12g0240900 expression was induced by jasmonic acid treatment in rice leaves prior to sakuranetin accumulation, and the Os12g0240900 protein showed reasonable kinetic properties to OsNOMT. On the basis of these results, we conclude that Os12g0240900 encodes an OsNOMT.
Figures





References
-
- Koga J., Shimura M., Oshima K., Ogawa N., Yamauchi T., Ogasawara N. (1995) Phytocassanes A, B, C, and D, novel diterpene phytoalexins from rice, Oryza sativa L. Tetrahedron 51, 7907–7918
-
- Koga J., Ogawa N., Yamauchi T., Kikuchi M., Ogasawara N., Shimura M. (1997) Functional moiety for the antifungal activity of phytocassane E, a diterpene phytoalexin from rice. Phytochemistry 44, 249–253
-
- Yajima A., Mori K. (2000) Synthesis and Absolute Configuration of (−)-Phytocassane D, a diterpene phytoalexin isolated from the rice plant, Oryza sativa. Eur. J. Org. Chem. 21, 4079–4091
-
- Akatsuka T., Kodama O., Sekido H., Kono Y., Takeuchi S. (1985) Novel phytoalexins (oryzalexins A, B, and C) isolated from rice blast leaves infected with Pyricularia oryzae. Part I: Isolation, characterization, and biological activities of oryzalexins Agric. Biol. Chem. 49, 1689–1694
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

