Development of a reverse-genetics system for murine norovirus 3: long-term persistence occurs in the caecum and colon
- PMID: 22495235
- PMCID: PMC3541523
- DOI: 10.1099/vir.0.042176-0
Development of a reverse-genetics system for murine norovirus 3: long-term persistence occurs in the caecum and colon
Abstract
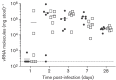
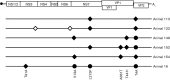
Human noroviruses (HuNoV) are a major cause of viral gastroenteritis worldwide, yet, due to the inability to propagate HuNoV in cell culture, murine norovirus (MNV) is typically used as a surrogate to study norovirus biology. MNV-3 represents an attractive strain to study norovirus infections in vivo because it establishes persistence in wild-type mice, yet causes symptoms resembling gastroenteritis in immune-compromised STAT1(-/-) mice. The lack of reverse-genetics approaches to recover genetically defined MNV-3 has limited further studies on the identification of viral sequences that contribute to persistence. Here we report the establishment of a combined DNA-based reverse-genetics and mouse-model system to study persistent MNV-3 infections in wild-type (C57BL/6) mice. Viral RNA and infectious virus were detected in faeces for at least 56 days after inoculation. Strikingly, the highest concentrations of viral RNA during persistence were detected in the caecum and colon, suggesting that viral persistence is maintained in these tissues. Possible adaptive changes arising during persistence in vivo appeared to accumulate in the minor capsid protein (VP2) and the viral polymerase (NS7), in contrast with adaptive mutations selected during cell-culture passages in RAW264.7 cells that appeared in the major capsid protein (VP1) and non-structural protein NS4. This system provides an attractive model that can be readily used to identify viral sequences that contribute to persistence in an immunocompetent host and to more acute infection in an immunocompromised host, providing new insights into the biology of norovirus infections.
Figures




References
-
- Bailey D., Karakasiliotis I., Vashist S., Chung L. M., Rees J., McFadden N., Benson A., Yarovinsky F., Simmonds P., Goodfellow I. (2010). Functional analysis of RNA structures present at the 3′ extremity of the murine norovirus genome: the variable polypyrimidine tract plays a role in viral virulence. J Virol 84, 2859–2870 10.1128/JVI.02053-09 - DOI - PMC - PubMed
-
- Barron E. L., Sosnovtsev S. V., Bok K., Prikhodko V., Sandoval-Jaime C., Rhodes C. R., Hasenkrug K., Carmody A. B., Ward J. M. & other authors (2011). Diversity of murine norovirus strains isolated from asymptomatic mice of different genetic backgrounds within a single U.S. research institute. PLoS ONE 6, e21435 10.1371/journal.pone.0021435 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

