In vivo regulation of the heme oxygenase-1 gene in humanized transgenic mice
- PMID: 22495295
- PMCID: PMC3396739
- DOI: 10.1038/ki.2012.102
In vivo regulation of the heme oxygenase-1 gene in humanized transgenic mice
Abstract
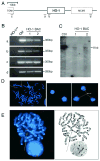
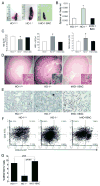
Heme oxygenase-1 (HO-1) catalyzes the rate-limiting step in heme degradation, producing equimolar amounts of carbon monoxide, iron, and biliverdin. Induction of HO-1 is a beneficial response to tissue injury in diverse animal models of diseases including acute kidney injury. In vitro analysis has shown that the human HO-1 gene is transcriptionally regulated by changes in chromatin conformation, but whether such control occurs in vivo is not known. To enable such an analysis, we generated transgenic mice, harboring an 87-kb bacterial artificial chromosome expressing human HO-1 mRNA and protein and bred these mice with HO-1 knockout mice to generate humanized BAC transgenic mice. This successfully rescued the phenotype of the knockout mice including reduced birth rates, tissue iron overload, splenomegaly, anemia, leukocytosis, dendritic cell abnormalities, and survival after acute kidney injury induced by rhabdomyolysis or cisplatin nephrotoxicity. Transcription factors such as USF1/2, JunB, Sp1, and CTCF were found to associate with regulatory regions of the human HO-1 gene in the kidney following rhabdomyolysis. Chromosome conformation capture and ChIP-loop assays confirmed this in the formation of chromatin looping in vivo. Thus, these bacterial artificial chromosome humanized HO-1 mice are a valuable model to study the human HO-1 gene, providing insight to the in vivo architecture of the gene in acute kidney injury and other diseases.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures




Comment in
-
Humanized HO-1 BAC transgenic mice: a new model for the study of HO-1 gene regulation.Kidney Int. 2012 Aug;82(3):253-5. doi: 10.1038/ki.2012.149. Kidney Int. 2012. PMID: 22791319
References
-
- Tenhunen R, Marver HS, Schmid R. Microsomal heme oxygenase. Characterization of the enzyme. J Biol Chem. 1969;244:6388–6394. - PubMed
-
- Maines MD. The heme oxygenase system: a regulator of second messenger gases. Annu Rev Pharmacol Toxicol. 1997;37:517–554. - PubMed
-
- Yoshida T, Kikuchi G. Purification and properties of heme oxygenase from pig spleen microsomes. J Biol Chem. 1978;253:4224–4229. - PubMed
-
- Hayashi S, Omata Y, Sakamoto H, et al. Characterization of rat heme oxygenase-3 gene. Implication of processed pseudogenes derived from heme oxygenase-2 gene. Gene. 2004;336:241–250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

