Genetic analysis of cytomegalovirus in malignant gliomas
- PMID: 22496213
- PMCID: PMC3393585
- DOI: 10.1128/JVI.00015-12
Genetic analysis of cytomegalovirus in malignant gliomas
Abstract
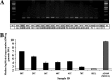
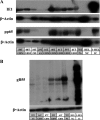
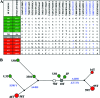
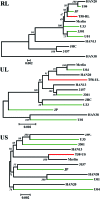
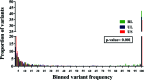
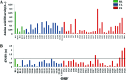
Human cytomegalovirus (HCMV) has been found in malignant gliomas at variable frequencies with efforts to date focused on characterizing the role(s) of single gene products in disease. Here, we reexamined the HCMV prevalence in malignant gliomas using different methods and began to dissect the genetics of HCMV in tumors. HCMV DNA was found in 16/17 (94%) tumor specimens. Viral DNA copy numbers were found to be low and variable, ranging from 10(2) to 10(6) copies/500 ng of total DNA. The tumor tissues had incongruences between viral DNA copy numbers and protein levels. However, nonlatent protein expression was detected in many tumors. The viral UL83 gene, encoding pp65, was found to segregate into five cancer-associated genotypes with a bias for amino acid changes in glioblastoma multiforme (GBM) in comparison to the low-grade tumors. Deep sequencing of a GBM-associated viral population resulted in 81,224 bp of genome coverage. Sequence analysis revealed the presence of intact open reading frames and higher numbers of high-frequency variations within the repeat long region compared to the unique long region, which harbors many core genes, and the unique short region (P = 0.001). This observation was in congruence with phylogenetic analyses across replication-competent viral strains in databases. The tumor-associated viral population was less variable (π = 0.1% and π(AA) = 0.08%) than that observed in other clinical infections. Moreover, 42/46 (91.3%) viral genes analyzed had dN/dS scores of <1, which is indicative of high amino acid sequence conservation. Taken together, these findings raise the possibility that replication-competent HCMV may exist in malignant gliomas.
Figures

References
-
- Bandelt H-J, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16:37–48 - PubMed
-
- Boccardo E, Villa LL. 2007. Viral origins of human cancer. Curr. Med. Chem. 14:2526–2539 - PubMed
-
- Cinatl J, Vogel JU, Kotchetkov R, Doerr WH. 2004. Oncomodulatory signals by regulatory proteins encoded by human cytomegalovirus: a novel role for viral infection in tumor progression. FEMS Microbiol. Rev. 28:59–77 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

