A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen
- PMID: 22496338
- PMCID: PMC3412965
- DOI: 10.1074/mcp.O111.013706
A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen
Abstract
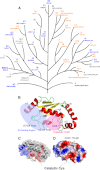
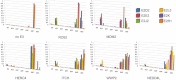
Here we describe a systematic structure-function analysis of the human ubiquitin (Ub) E2 conjugating proteins, consisting of the determination of 15 new high-resolution three-dimensional structures of E2 catalytic domains, and autoubiquitylation assays for 26 Ub-loading E2s screened against a panel of nine different HECT (homologous to E6-AP carboxyl terminus) E3 ligase domains. Integration of our structural and biochemical data revealed several E2 surface properties associated with Ub chain building activity; (1) net positive or neutral E2 charge, (2) an "acidic trough" located near the catalytic Cys, surrounded by an extensive basic region, and (3) similarity to the previously described HECT binding signature in UBE2L3 (UbcH7). Mass spectrometry was used to characterize the autoubiquitylation products of a number of functional E2-HECT pairs, and demonstrated that HECT domains from different subfamilies catalyze the formation of very different types of Ub chains, largely independent of the E2 in the reaction. Our data set represents the first comprehensive analysis of E2-HECT E3 interactions, and thus provides a framework for better understanding the molecular mechanisms of ubiquitylation.
Figures



References
-
- Schwartz A. L., Ciechanover A. (2009) Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu. Rev. Pharmacol. Toxicol. 49, 73–96 - PubMed
-
- Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 - PubMed
-
- Pickart C. M., Eddins M. J. (2004) Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 - PubMed
-
- Pickart C. M., Fushman D. (2004) Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

