TFIIIC localizes budding yeast ETC sites to the nuclear periphery
- PMID: 22496415
- PMCID: PMC3395662
- DOI: 10.1091/mbc.E11-04-0365
TFIIIC localizes budding yeast ETC sites to the nuclear periphery
Abstract
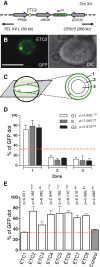
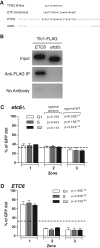
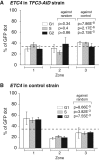
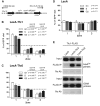
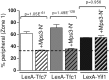
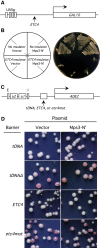
Chromatin function requires specific three-dimensional architectures of chromosomes. We investigated whether Saccharomyces cerevisiae extra TFIIIC (ETC) sites, which bind the TFIIIC transcription factor but do not recruit RNA polymerase III, show specific intranuclear positioning. We show that six of the eight known S. cerevisiae ETC sites localize predominantly at the nuclear periphery, and that ETC sites retain their tethering function when moved to a new chromosomal location. Several lines of evidence indicate that TFIIIC is central to the ETC peripheral localization mechanism. Mutating or deleting the TFIIIC-binding consensus ablated ETC -site peripheral positioning, and inducing degradation of the TFIIIC subunit Tfc3 led to rapid release of an ETC site from the nuclear periphery. We find, moreover, that anchoring one TFIIIC subunit at an ectopic chromosomal site causes recruitment of others and drives peripheral tethering. Localization of ETC sites at the nuclear periphery also requires Mps3, a Sad1-UNC-84-domain protein that spans the inner nuclear membrane. Surprisingly, we find that the chromatin barrier and insulator functions of an ETC site do not depend on correct peripheral localization. In summary, TFIIIC and Mps3 together direct the intranuclear positioning of a new class of S. cerevisiae genomic loci positioned at the nuclear periphery.
Figures



References
-
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. - PubMed
-
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. - PubMed
-
- Baker RE, Gabrielsen O, Hall BD. Effects of tRNATyr point mutations on the binding of yeast RNA polymerase III transcription factor C. J Biol Chem. 1986;261:5275–5282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

