An interdomain binding site on HIV-1 Nef interacts with PACS-1 and PACS-2 on endosomes to down-regulate MHC-I
- PMID: 22496420
- PMCID: PMC3364181
- DOI: 10.1091/mbc.E11-11-0928
An interdomain binding site on HIV-1 Nef interacts with PACS-1 and PACS-2 on endosomes to down-regulate MHC-I
Abstract
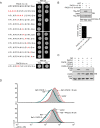
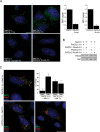
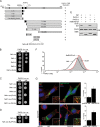
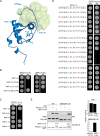
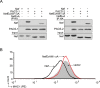
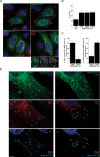
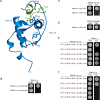
The human immunodeficiency virus type 1 (HIV-1) accessory protein Nef directs virus escape from immune surveillance by subverting host cell intracellular signaling and membrane traffic to down-regulate cell-surface major histocompatibility complex class I (MHC-I). The interaction of Nef with the sorting proteins PACS-1 and PACS-2 mediates key signaling and trafficking steps required for Nef-mediated MHC-I down-regulation. Little is known, however, about the molecular basis underlying the Nef-PACS interaction. Here we identify the sites on Nef and the PACS proteins required for their interaction and describe the consequences of disrupting this interaction for Nef action. A previously unidentified cargo subsite on PACS-1 and PACS-2 interacted with a bipartite site on Nef formed by the EEEE(65) acidic cluster on the N-terminal domain and W(113) in the core domain. Mutation of these sites prevented the interaction between Nef and the PACS proteins on Rab5 (PACS-2 and PACS-1)- or Rab7 (PACS-1)-positive endosomes as determined by bimolecular fluorescence complementation and caused a Nef mutant defective in PACS binding to localize to distorted endosomal compartments. Consequently, disruption of the Nef-PACS interaction repressed Nef-induced MHC-I down-regulation in peripheral blood mononuclear cells. Our results provide insight into the molecular basis of Nef action and suggest new strategies to combat HIV-1.
Figures

References
-
- Arold S, Franken P, Strub MP, Hoh F, Benichou S, Benarous R, Dumas C. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure. 1997;5:1361–1372. - PubMed
-
- Atkins KM, Thomas L, Youker RT, Harriff MJ, Pissani F, You H, Thomas G. HIV-1 Nef binds PACS-2 to assemble a multikinase cascade that triggers major histocompatibility complex class I (MHC-I) down-regulation: analysis using short interfering RNA and knock-out mice. J Biol Chem. 2008;283:11772–11784. - PMC - PubMed
-
- Blagoveshchenskaya AD, Thomas L, Feliciangeli SF, Hung CH, Thomas G. HIV-1 Nef down-regulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell. 2002;111:853–866. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

