Identification of a conserved interface between PUF and CPEB proteins
- PMID: 22496444
- PMCID: PMC3365739
- DOI: 10.1074/jbc.M112.352815
Identification of a conserved interface between PUF and CPEB proteins
Abstract
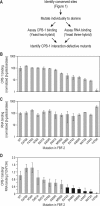
Members of the PUF (Pumilio and FBF) and CPEB (cytoplasmic polyadenylation element-binding) protein families collaborate to regulate mRNA expression throughout eukaryotes. Here, we focus on the physical interactions between members of these two families, concentrating on Caenorhabditis elegans FBF-2 and CPB-1. To localize the site of interaction on FBF-2, we identified conserved amino acids within C. elegans PUF proteins. Deletion of an extended loop containing several conserved residues abolished binding to CPB-1. We analyzed alanine substitutions at 13 individual amino acids in FBF-2, each identified via its conservation. Multiple single point mutations disrupted binding to CPB-1 but not to RNA. Position Tyr-479 was particularly critical as multiple substitutions to other amino acids at this position did not restore binding. The complex of FBF-2 and CPB-1 repressed translation of an mRNA containing an FBF binding element. Repression required both proteins and was disrupted by FBF-2 alleles that failed to bind CPB-1 or RNA. The equivalent loop in human PUM2 is required for binding to human CPEB3 in vitro, although the primary sequences of the human and C. elegans PUF proteins have diverged in that region. Our findings define a key region in PUF/CPEB interactions and imply a conserved platform through which PUF proteins interact with their protein partners.
Figures




References
-
- Valencia-Sanchez M. A., Liu J., Hannon G. J., Parker R. (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20, 515–524 - PubMed
-
- Besse F., Ephrussi A. (2008) Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat. Rev. Mol. Cell Biol. 9, 971–980 - PubMed
-
- Osborne H. B., Richter J. D. (1997) Translational control by polyadenylation during early development. Prog. Mol. Subcell. Biol. 18, 173–198 - PubMed
-
- Wickens M., Kimble J., Strickland S. (1996) Translational control of developmental decisions in Translational Control (Hershey J., Mathews M., Sonenberg N., eds) pp. 411–450, Cold Spring Harbor Laboratory Press, Plainview, NY
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

