Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces death receptor 5 networks that are highly organized
- PMID: 22496450
- PMCID: PMC3375548
- DOI: 10.1074/jbc.M111.306480
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces death receptor 5 networks that are highly organized
Abstract
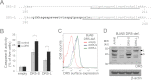
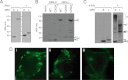
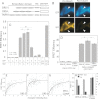
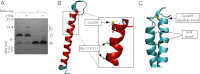
Recent evidence suggests that TNF-related apoptosis-inducing ligand (TRAIL), a death-inducing cytokine with anti-tumor potential, initiates apoptosis by re-organizing TRAIL receptors into large clusters, although the structure of these clusters and the mechanism by which they assemble are unknown. Here, we demonstrate that TRAIL receptor 2 (DR5) forms receptor dimers in a ligand-dependent manner at endogenous receptor levels, and these receptor dimers exist within high molecular weight networks. Using mutational analysis, FRET, fluorescence microscopy, synthetic biochemistry, and molecular modeling, we find that receptor dimerization relies upon covalent and noncovalent interactions between membrane-proximal residues. Additionally, by using FRET, we show that the oligomeric structure of two functional isoforms of DR5 is indistinguishable. The resulting model of DR5 activation should revise the accepted architecture of the functioning units of DR5 and the structurally homologous TNF receptor superfamily members.
Figures


References
-
- Wiley S. R., Schooley K., Smolak P. J., Din W. S., Huang C. P., Nicholl J. K., Sutherland G. R., Smith T. D., Rauch C., Smith C. A. (1995) Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 3, 673–682 - PubMed
-
- Pan G., Ni J., Wei Y. F., Yu G., Gentz R., Dixit V. M. (1997) An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 277, 815–818 - PubMed
-
- Pan G., O'Rourke K., Chinnaiyan A. M., Gentz R., Ebner R., Ni J., Dixit V. M. (1997) The receptor for the cytotoxic ligand TRAIL. Science 276, 111–113 - PubMed
-
- Ashkenazi A., Holland P., Eckhardt S. G. (2008) Ligand-based targeting of apoptosis in cancer. The potential of recombinant human apoptosis ligand 2/tumor necrosis factor-related apoptosis-inducing ligand (rhApo2L/TRAIL). J. Clin. Oncol. 26, 3621–3630 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

