IκB kinases modulate the activity of the androgen receptor in prostate carcinoma cell lines
- PMID: 22496618
- PMCID: PMC3323896
- DOI: 10.1593/neo.111444
IκB kinases modulate the activity of the androgen receptor in prostate carcinoma cell lines
Abstract
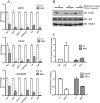
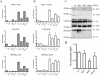
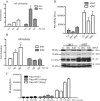
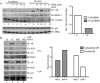
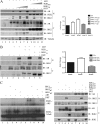
Enhanced nuclear localization of nuclear factor κB (NF-κB) in prostate cancer (PCa) samples and constitutive NF-κB signaling in a class of PCa cell lines with low androgen receptor (AR) expression (PC3 and DU-145) imply an important role of the IκB kinase (IKK)/NF-κB system in PCa. However, most PCa and PCa cell lines depend on the activity of the AR, and the role of NF-κB in these AR-expressing PCa remains unclear. Here, we demonstrate that inhibition of NF-κB signaling by the IKK inhibitor BMS345541 reduced proliferation and increased apoptosis in AR-expressing PCa cell lines. Furthermore, AR activity and target gene expression were distinctively reduced, whereas AR protein levels remained unaltered on BMS345541 treatment. Similar effects were observed particularly after small interfering RNA (siRNA)-mediated knockdown of IKK1, but not by siRNA-mediated suppression of IKK2. Moreover, IKK1 overexpression augmented 5α-dihydrotestosterone-induced nuclear AR translocation, whereas nuclear AR was reduced by IKK1 knockdown or BMS345541. However, because IKK1 also enhances the activity of a chronically nuclear AR mutant, modulation of the subcellular distribution seems not to be the only mechanism by which IKK1 enhances AR activity. Finally, reduced in vivo AR phosphorylation after BMS345541 treatment and in vitro AR phosphorylation by IKK1 or IKK2 imply that AR constitutes a novel IKK target. Taken together, our data identify IKK1 as a potentially target structure for future therapeutic intervention in PCa.
Copyright © 2012 Neoplasia Press, Inc.
Figures

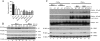

References
-
- Linja MJ, Savinainen KJ, Saramaki OR, Tammela TL, Vessella RL, Visakorpi T. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001;61:3550–3555. - PubMed
-
- Zhao XY, Malloy PJ, Krishnan AV, Swami S, Navone NM, Peehl DM, Feldman D. Glucocorticoids can promote androgen-independent growth of prostate cancer cells through a mutated androgen receptor. Nat Med. 2000;6:703–706. - PubMed
-
- Scheidereit C. IκB kinase complexes: gateways to NF-κB activation and transcription. Oncogene. 2006;25:6685–6705. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
