Improved statistics for genome-wide interaction analysis
- PMID: 22496670
- PMCID: PMC3320596
- DOI: 10.1371/journal.pgen.1002625
Improved statistics for genome-wide interaction analysis
Abstract
Recently, Wu and colleagues [1] proposed two novel statistics for genome-wide interaction analysis using case/control or case-only data. In computer simulations, their proposed case/control statistic outperformed competing approaches, including the fast-epistasis option in PLINK and logistic regression analysis under the correct model; however, reasons for its superior performance were not fully explored. Here we investigate the theoretical properties and performance of Wu et al.'s proposed statistics and explain why, in some circumstances, they outperform competing approaches. Unfortunately, we find minor errors in the formulae for their statistics, resulting in tests that have higher than nominal type 1 error. We also find minor errors in PLINK's fast-epistasis and case-only statistics, although theory and simulations suggest that these errors have only negligible effect on type 1 error. We propose adjusted versions of all four statistics that, both theoretically and in computer simulations, maintain correct type 1 error rates under the null hypothesis. We also investigate statistics based on correlation coefficients that maintain similar control of type 1 error. Although designed to test specifically for interaction, we show that some of these previously-proposed statistics can, in fact, be sensitive to main effects at one or both loci, particularly in the presence of linkage disequilibrium. We propose two new "joint effects" statistics that, provided the disease is rare, are sensitive only to genuine interaction effects. In computer simulations we find, in most situations considered, that highest power is achieved by analysis under the correct genetic model. Such an analysis is unachievable in practice, as we do not know this model. However, generally high power over a wide range of scenarios is exhibited by our joint effects and adjusted Wu statistics. We recommend use of these alternative or adjusted statistics and urge caution when using Wu et al.'s originally-proposed statistics, on account of the inflated error rate that can result.
© 2012 Ueki, Cordell.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
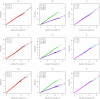
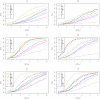
 . Top panels ((a) and (b)): Case/Control not in LD; Middle panels ((c) and (d)): Case/Control in LD; Bottom panels ((e) and (f)): Case-Only not in LD; Left hand panels ((a), (c) and (e)): No main effect; Right hand panels ((b), (d) and (f)): Locus G has main effect; FE: Fast-Epistasis; AFE: Adjusted FE; Wu: Wu et al. statistic; AWu: Adjusted Wu statistic; WZ: Wellek and Ziegler statistic; JE: Joint Effects statistic; IWu: Ideal Wu statistic; C: Logistic regression using correct coding; IC: Logistic regression using incorrect ( = Recessive
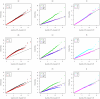
. Top panels ((a) and (b)): Case/Control not in LD; Middle panels ((c) and (d)): Case/Control in LD; Bottom panels ((e) and (f)): Case-Only not in LD; Left hand panels ((a), (c) and (e)): No main effect; Right hand panels ((b), (d) and (f)): Locus G has main effect; FE: Fast-Epistasis; AFE: Adjusted FE; Wu: Wu et al. statistic; AWu: Adjusted Wu statistic; WZ: Wellek and Ziegler statistic; JE: Joint Effects statistic; IWu: Ideal Wu statistic; C: Logistic regression using correct coding; IC: Logistic regression using incorrect ( = Recessive Dominant) coding; WZC: Wellek and Ziegler case-only statistic using correct coding; WZIC: Wellek and Ziegler case-only statistic using incorrect ( = Recessive
Dominant) coding; WZC: Wellek and Ziegler case-only statistic using correct coding; WZIC: Wellek and Ziegler case-only statistic using incorrect ( = Recessive Dominant) coding.
Dominant) coding.
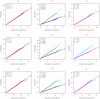
 . Top panels ((a) and (b)): Case/Control not in LD; Middle panels ((c) and (d)): Case/Control in LD; Bottom panels ((e) and (f)): Case-Only not in LD; Left hand panels ((a), (c) and (e)): No main effect; Right hand panels ((b), (d) and (f)): Locus G has main effect; FE: Fast-Epistasis; AFE: Adjusted FE; Wu: Wu et al. statistic; AWu: Adjusted Wu statistic; WZ: Wellek and Ziegler statistic; JE: Joint Effects statistic; IWu: Ideal Wu statistic; C: Logistic regression using correct coding; IC: Logistic regression using incorrect ( = Dominant
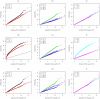
. Top panels ((a) and (b)): Case/Control not in LD; Middle panels ((c) and (d)): Case/Control in LD; Bottom panels ((e) and (f)): Case-Only not in LD; Left hand panels ((a), (c) and (e)): No main effect; Right hand panels ((b), (d) and (f)): Locus G has main effect; FE: Fast-Epistasis; AFE: Adjusted FE; Wu: Wu et al. statistic; AWu: Adjusted Wu statistic; WZ: Wellek and Ziegler statistic; JE: Joint Effects statistic; IWu: Ideal Wu statistic; C: Logistic regression using correct coding; IC: Logistic regression using incorrect ( = Dominant Recessive) coding; WZC: Wellek and Ziegler case-only statistic using correct coding; WZIC: Wellek and Ziegler case-only statistic using incorrect ( = Dominant
Recessive) coding; WZC: Wellek and Ziegler case-only statistic using correct coding; WZIC: Wellek and Ziegler case-only statistic using incorrect ( = Dominant Recessive) coding.
Recessive) coding.Comment on
-
A novel statistic for genome-wide interaction analysis.PLoS Genet. 2010 Sep 23;6(9):e1001131. doi: 10.1371/journal.pgen.1001131. PLoS Genet. 2010. PMID: 20885795 Free PMC article.
References
-
- Wu X, Dong H, Luo L, Zhu Y, Peng G, et al. A novel statistic for genome-wide interaction analysis. PLoS Genet. 2010;6:e1001131. doi: 10.1371/journal.pgen.1001131. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

