Complete chloroplast genome sequence of an orchid model plant candidate: Erycina pusilla apply in tropical Oncidium breeding
- PMID: 22496851
- PMCID: PMC3319614
- DOI: 10.1371/journal.pone.0034738
Complete chloroplast genome sequence of an orchid model plant candidate: Erycina pusilla apply in tropical Oncidium breeding
Abstract
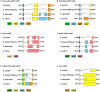
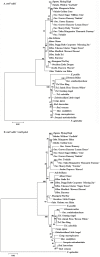
Oncidium is an important ornamental plant but the study of its functional genomics is difficult. Erycina pusilla is a fast-growing Oncidiinae species. Several characteristics including low chromosome number, small genome size, short growth period, and its ability to complete its life cycle in vitro make E. pusilla a good model candidate and parent for hybridization for orchids. Although genetic information remains limited, systematic molecular analysis of its chloroplast genome might provide useful genetic information. By combining bacterial artificial chromosome (BAC) clones and next-generation sequencing (NGS), the chloroplast (cp) genome of E. pusilla was sequenced accurately, efficiently and economically. The cp genome of E. pusilla shares 89 and 84% similarity with Oncidium Gower Ramsey and Phalanopsis aphrodite, respectively. Comparing these 3 cp genomes, 5 regions have been identified as showing diversity. Using PCR analysis of 19 species belonging to the Epidendroideae subfamily, a conserved deletion was found in the rps15-trnN region of the Cymbidieae tribe. Because commercial Oncidium varieties in Taiwan are limited, identification of potential parents using molecular breeding method has become very important. To demonstrate the relationship between taxonomic position and hybrid compatibility of E. pusilla, 4 DNA regions of 36 tropically adapted Oncidiinae varieties have been analyzed. The results indicated that trnF-ndhJ and trnH-psbA were suitable for phylogenetic analysis. E. pusilla proved to be phylogenetically closer to Rodriguezia and Tolumnia than Oncidium, despite its similar floral appearance to Oncidium. These results indicate the hybrid compatibility of E. pusilla, its cp genome providing important information for Oncidium breeding.
Conflict of interest statement
Figures

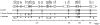

References
-
- Chase MW, Cameron KM, Barrett RL, Freudenstein JV. DNA data and Orchidaceae systematics: a new phylogenetic classification. In: Dixon KWKS, Barrett RL, Cribb PJ, editors. Orchid conservation. Kota Kinabalu: Natural History Publications (Borneo); 2003. pp. 69–89.
-
- Kerbauy GB. In vitro flowering of Oncidium varicosum mericlones (Orchidaceae). Plant Sci Lett. 1984;35:73–75.
-
- Chen CC, Hsu YM. Effect of accumulation temperature on the flower quality of Oncidium. J Agric For. 2003;52:33–48 (in Chinese).
-
- Hsiao YY, Pan ZJ, Hsu CC, Yang YP, Hsu YC, et al. Research on orchid biology and biotechnology. Plant Cell Physiol. 2011;52:1467–1486. - PubMed
-
- Fu CH, Chen YW, Hsiao YY, Pan ZJ, Liu ZJ, et al. OrchidBase: a collection of sequences of the transcriptome derived from orchids. Plant Cell Physiol. 2011;52:238–243. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

