Binding affects the tertiary and quaternary structures of the Shigella translocator protein IpaB and its chaperone IpgC
- PMID: 22497344
- PMCID: PMC3903306
- DOI: 10.1021/bi300243z
Binding affects the tertiary and quaternary structures of the Shigella translocator protein IpaB and its chaperone IpgC
Abstract
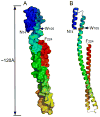
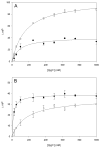
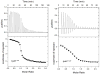
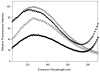
Shigella flexneri uses its type III secretion system (T3SS) to promote invasion of human intestinal epithelial cells as the first step in causing shigellosis, a life-threatening form of dysentery. The Shigella type III secretion apparatus (T3SA) consists of a basal body that spans the bacterial envelope and an exposed needle that injects effector proteins into target cells. The nascent Shigella T3SA needle is topped with a pentamer of the needle tip protein invasion plasmid antigen D (IpaD). Bile salts trigger recruitment of the first hydrophobic translocator protein, IpaB, to the tip complex where it senses contact with a host membrane. In the bacterial cytoplasm, IpaB exists in a complex with its chaperone IpgC. Several structures of IpgC have been determined, and we recently reported the 2.1 Å crystal structure of the N-terminal domain (IpaB(74.224)) of IpaB. Like IpgC, the IpaB N-terminal domain exists as a homodimer in solution. We now report that when the two are mixed, these homodimers dissociate and form heterodimers having a nanomolar dissociation constant. This is consistent with the equivalent complexes copurified after they had been co-expressed in Escherichia coli. Fluorescence data presented here also indicate that the N-terminal domain of IpaB possesses two regions that appear to contribute additively to chaperone binding. It is also likely that the N-terminus of IpaB adopts an alternative conformation as a result of chaperone binding. The importance of these findings within the functional context of these proteins is discussed.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

