Multi-source feature learning for joint analysis of incomplete multiple heterogeneous neuroimaging data
- PMID: 22498655
- PMCID: PMC3358419
- DOI: 10.1016/j.neuroimage.2012.03.059
Multi-source feature learning for joint analysis of incomplete multiple heterogeneous neuroimaging data
Abstract
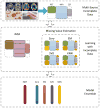
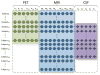
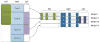
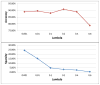
Analysis of incomplete data is a big challenge when integrating large-scale brain imaging datasets from different imaging modalities. In the Alzheimer's Disease Neuroimaging Initiative (ADNI), for example, over half of the subjects lack cerebrospinal fluid (CSF) measurements; an independent half of the subjects do not have fluorodeoxyglucose positron emission tomography (FDG-PET) scans; many lack proteomics measurements. Traditionally, subjects with missing measures are discarded, resulting in a severe loss of available information. In this paper, we address this problem by proposing an incomplete Multi-Source Feature (iMSF) learning method where all the samples (with at least one available data source) can be used. To illustrate the proposed approach, we classify patients from the ADNI study into groups with Alzheimer's disease (AD), mild cognitive impairment (MCI) and normal controls, based on the multi-modality data. At baseline, ADNI's 780 participants (172AD, 397 MCI, 211 NC), have at least one of four data types: magnetic resonance imaging (MRI), FDG-PET, CSF and proteomics. These data are used to test our algorithm. Depending on the problem being solved, we divide our samples according to the availability of data sources, and we learn shared sets of features with state-of-the-art sparse learning methods. To build a practical and robust system, we construct a classifier ensemble by combining our method with four other methods for missing value estimation. Comprehensive experiments with various parameters show that our proposed iMSF method and the ensemble model yield stable and promising results.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

References
-
- 2011 Alzheimer’s Disease Facts and Figures. http://www.alz.org.
-
- Ando RK, Zhang T. A framework for learning predictive structures from multiple tasks and unlabeled data. The Journal of Machine Learning Research. 2005;6:1817–1853.
-
- Argyriou A, Evgeniou T, Pontil M. Convex multi-task feature learning. Machine Learning. 2008;73:243–272.
-
- Ashburner J, Friston K. Multimodal image coregistration and partitioning--a unified framework. Neuroimage. 1997;6:209–217. - PubMed
-
- Braskie MN, Klunder AD, Hayashi KM, Protas H, Kepe V, Miller KJ, Huang SC, Barrio JR, Ercoli LM, Siddarth P, Satyamurthy N, Liu J, Toga AW, Bookheimer SY, Small GW, Thompson PM. Plaque and tangle imaging and cognition in normal aging and Alzheimer’s disease. Neurobiol Aging. 2008;31:1669–1678. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AG016570/AG/NIA NIH HHS/United States
- R01 LM010730/LM/NLM NIH HHS/United States
- R01 AG040060/AG/NIA NIH HHS/United States
- EB01651/EB/NIBIB NIH HHS/United States
- P30 AG013846/AG/NIA NIH HHS/United States
- LM05639/LM/NLM NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- K01 AG030514/AG/NIA NIH HHS/United States
- R21 EB001561/EB/NIBIB NIH HHS/United States
- R21 RR019771/RR/NCRR NIH HHS/United States
- R01 LM005639/LM/NLM NIH HHS/United States
- U01 AG024904/AG/NIA NIH HHS/United States
- U19 AG010483/AG/NIA NIH HHS/United States
- RR019771/RR/NCRR NIH HHS/United States
- P50 AG016570/AG/NIA NIH HHS/United States
- R01 MH097268/MH/NIMH NIH HHS/United States
- P30 AG010129/AG/NIA NIH HHS/United States

