Digital clocks: simple Boolean models can quantitatively describe circadian systems
- PMID: 22499125
- PMCID: PMC3405750
- DOI: 10.1098/rsif.2012.0080
Digital clocks: simple Boolean models can quantitatively describe circadian systems
Erratum in
- J R Soc Interface. 2012 Dec 7;9(77):3578
Abstract
The gene networks that comprise the circadian clock modulate biological function across a range of scales, from gene expression to performance and adaptive behaviour. The clock functions by generating endogenous rhythms that can be entrained to the external 24-h day-night cycle, enabling organisms to optimally time biochemical processes relative to dawn and dusk. In recent years, computational models based on differential equations have become useful tools for dissecting and quantifying the complex regulatory relationships underlying the clock's oscillatory dynamics. However, optimizing the large parameter sets characteristic of these models places intense demands on both computational and experimental resources, limiting the scope of in silico studies. Here, we develop an approach based on Boolean logic that dramatically reduces the parametrization, making the state and parameter spaces finite and tractable. We introduce efficient methods for fitting Boolean models to molecular data, successfully demonstrating their application to synthetic time courses generated by a number of established clock models, as well as experimental expression levels measured using luciferase imaging. Our results indicate that despite their relative simplicity, logic models can (i) simulate circadian oscillations with the correct, experimentally observed phase relationships among genes and (ii) flexibly entrain to light stimuli, reproducing the complex responses to variations in daylength generated by more detailed differential equation formulations. Our work also demonstrates that logic models have sufficient predictive power to identify optimal regulatory structures from experimental data. By presenting the first Boolean models of circadian circuits together with general techniques for their optimization, we hope to establish a new framework for the systematic modelling of more complex clocks, as well as other circuits with different qualitative dynamics. In particular, we anticipate that the ability of logic models to provide a computationally efficient representation of system behaviour could greatly facilitate the reverse-engineering of large-scale biochemical networks.
Figures
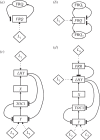
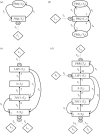
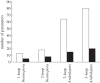
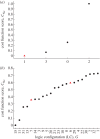

References
-
- Dunlap J. C., Loros J. J., DeCoursey P. J. 2003. Chronobiology: biological timekeeping. Sunderland, MA: Sinauer Associates
-
- Young M. W., Kay S. A. 2001. Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2, 702–71510.1038/35088576 (doi:10.1038/35088576) - DOI - DOI - PubMed
-
- Bell-Pedersen D., Cassone V. M., Earnest D. J., Golden S. S., Hardin P. E., Thomas T. L., Zoran M. 2005. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 6, 544–55610.1038/nrg1633 (doi:10.1038/nrg1633) - DOI - DOI - PMC - PubMed
-
- Zhang E. E., Kay S. A. 2010. Clocks not winding down: unravelling circadian networks. Nat. Rev. Mol. Cell Biol. 11, 764–77610.1038/nrm2995 (doi:10.1038/nrm2995) - DOI - DOI - PubMed
-
- Khapre R. V., Samsa W. E., Kondatov R. V. 2010. Circadian regulation of cell cycle: molecular connections between aging and the circadian clock. Ann. Med. 42, 1695–170010.3109/07853890.2010.499134 (doi:10.3109/07853890.2010.499134) - DOI - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
