Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions
- PMID: 22499787
- PMCID: PMC3358852
- DOI: 10.1073/pnas.1200566109
Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions
Abstract
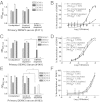
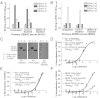
Dengue is a mosquito-borne flavivirus that is spreading at an unprecedented rate and has developed into a major health and economic burden in over 50 countries. Even though infected individuals develop potent and long-lasting serotype-specific neutralizing antibodies (Abs), the epitopes engaged by human neutralizing Abs have not been identified. Here, we demonstrate that the dengue virus (DENV)-specific serum Ab response in humans consists of a large fraction of cross-reactive, poorly neutralizing Abs and a small fraction of serotype-specific, potently inhibitory Abs. Although many mouse-generated, strongly neutralizing monoclonal antibodies (mAbs) recognize epitopes that are present on recombinant DENV envelope (E) proteins, unexpectedly, the majority of neutralizing Abs in human immune sera bound to intact virions but not to the ectodomain of purified soluble E proteins. These conclusions with polyclonal Abs were confirmed with newly generated human mAbs derived from DENV-immune individuals. Two of three strongly neutralizing human mAbs bound to E protein epitopes that were preserved on the virion but not on recombinant E (rE) protein. We propose that humans produce Abs that neutralize DENV infection by binding a complex, quaternary structure epitope that is expressed only when E proteins are assembled on a virus particle. Mapping studies indicate that this epitope has a footprint that spans adjacent E protein dimers and includes residues at the hinge between domains I and II of E protein. These results have significant implications for the DENV Ab and vaccine field.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: The spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10(Suppl):S98–S109. - PubMed
-
- Beasley DWC, Barrett ADT. The Infectious Agent, Dengue, Tropical Medicine: Science and Practice. In: Halstead SBP, Hoffman G, editors. Vol 5. London: Imperial College Press; 2008. pp. 29–73.
-
- Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res. 2003;60:421–467. - PubMed
-
- Roehrig JT. Antigenic structure of flavivirus proteins. Adv Virus Res. 2003;59:141–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

