Using gene expression noise to understand gene regulation
- PMID: 22499939
- PMCID: PMC3358231
- DOI: 10.1126/science.1216379
Using gene expression noise to understand gene regulation
Abstract
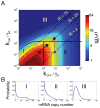
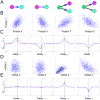
Phenotypic variation is ubiquitous in biology and is often traceable to underlying genetic and environmental variation. However, even genetically identical cells in identical environments display variable phenotypes. Stochastic gene expression, or gene expression "noise," has been suggested as a major source of this variability, and its physiological consequences have been topics of intense research for the last decade. Several recent studies have measured variability in protein and messenger RNA levels, and they have discovered strong connections between noise and gene regulation mechanisms. When integrated with discrete stochastic models, measurements of cell-to-cell variability provide a sensitive "fingerprint" with which to explore fundamental questions of gene regulation. In this review, we highlight several studies that used gene expression variability to develop a quantitative understanding of the mechanisms and dynamics of gene regulation.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

