miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay
- PMID: 22499947
- PMCID: PMC3971879
- DOI: 10.1126/science.1215691
miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay
Abstract
microRNAs (miRNAs) regulate gene expression through translational repression and/or messenger RNA (mRNA) deadenylation and decay. Because translation, deadenylation, and decay are closely linked processes, it is important to establish their ordering and thus to define the molecular mechanism of silencing. We have investigated the kinetics of these events in miRNA-mediated gene silencing by using a Drosophila S2 cell-based controllable expression system and show that mRNAs with both natural and engineered 3' untranslated regions with miRNA target sites are first subject to translational inhibition, followed by effects on deadenylation and decay. We next used a natural translational elongation stall to show that miRNA-mediated silencing inhibits translation at an early step, potentially translation initiation.
Figures

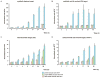
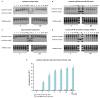

Comment in
-
Small RNAs: miRNAs' strict schedule.Nat Rev Genet. 2012 May 3;13(6):378. doi: 10.1038/nrg3251. Nat Rev Genet. 2012. PMID: 22552258 No abstract available.
-
miRNAs' strict schedule.Nat Rev Mol Cell Biol. 2012 May 3;13(6):340-1. doi: 10.1038/nrm3354. Nat Rev Mol Cell Biol. 2012. PMID: 22552328 No abstract available.
References
-
- Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. - PubMed
-
- Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. - PubMed
-
- Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

