Methyl effects on protein-ligand binding
- PMID: 22500930
- PMCID: PMC3353327
- DOI: 10.1021/jm3003697
Methyl effects on protein-ligand binding
Abstract
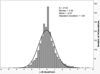
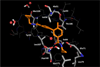
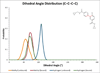
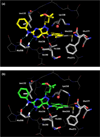
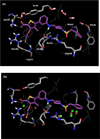
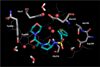
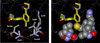
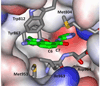
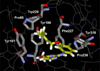
The effects of addition of a methyl group to a lead compound on biological activity are examined. A literature analysis of >2000 cases reveals that an activity boost of a factor of 10 or more is found with an 8% frequency, and a 100-fold boost is a 1 in 200 event. Four cases in the latter category are analyzed in depth to elucidate any unusual aspects of the protein-ligand binding, distribution of water molecules, and changes in conformational energetics. The analyses include Monte Carlo/free-energy perturbation (MC/FEP) calculations for methyl replacements in inhibitor series for p38α MAP kinase, ACK1, PTP1B, and thrombin. Methyl substitutions ortho to an aryl ring can be particularly effective at improving activity by inducing a propitious conformational change. The greatest improvements in activity arise from coupling the conformational gain with the burial of the methyl group in a hydrophobic region of the protein.
Figures
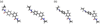


References
-
- Barreiro EJ, Kümmerle AE, Fraga CAM. The Methylation Effect in Medicinal Chemistry. Chem. Rev. 2011;111:5215–5246. - PubMed
-
- Jorgensen WL. Free Energy Calculations, a Breakthrough for Modeling Organic Chemistry in Solution. Acc. Chem. Res. 1989;22:184–189.
-
- Kollman P. Free Energy Calculations: Applications to Chemical and Biochemical Phenomena. Chem. Rev. 1993;93:2395–2417.
-
- Lamb ML, Jorgensen WL. Curr. Opin. Chem. Biol. 1997;1:449–457. - PubMed
-
- Simonson T, Georgios A, Karplus M. Free Energy Simulations Come of Age: Protein-Ligand Recognition. Acc. Chem. Res. 2002;35:430–437. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

