Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids
- PMID: 22503634
- PMCID: PMC3376635
- DOI: 10.1016/j.ajhg.2012.03.014
Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids
Abstract
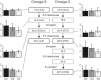
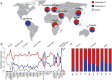
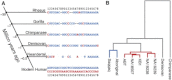
Omega-3 and omega-6 long-chain polyunsaturated fatty acids (LC-PUFAs) are essential for the development and function of the human brain. They can be obtained directly from food, e.g., fish, or synthesized from precursor molecules found in vegetable oils. To determine the importance of genetic variability to fatty-acid biosynthesis, we studied FADS1 and FADS2, which encode rate-limiting enzymes for fatty-acid conversion. We performed genome-wide genotyping (n = 5,652 individuals) and targeted resequencing (n = 960 individuals) of the FADS region in five European population cohorts. We also analyzed available genomic data from human populations, archaic hominins, and more distant primates. Our results show that present-day humans have two common FADS haplotypes-defined by 28 closely linked SNPs across 38.9 kb-that differ dramatically in their ability to generate LC-PUFAs. No independent effects on FADS activity were seen for rare SNPs detected by targeted resequencing. The more efficient, evolutionarily derived haplotype appeared after the lineage split leading to modern humans and Neanderthals and shows evidence of positive selection. This human-specific haplotype increases the efficiency of synthesizing essential long-chain fatty acids from precursors and thereby might have provided an advantage in environments with limited access to dietary LC-PUFAs. In the modern world, this haplotype has been associated with lifestyle-related diseases, such as coronary artery disease.
Copyright © 2012 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Leonard W.R., Snodgrass J.J., Robertson M.L. Evolutionary Perspectives on Fat Ingestion and Metabolism in Humans. In: Montmighteur J.P., le Coutre J., editors. Fat Detection: Taste, Texture, and Post Ingestive Effects. CRC Press; Boca Raton, FL: 2010. - PubMed
-
- Darios F., Davletov B. Omega-3 and omega-6 fatty acids stimulate cell membrane expansion by acting on syntaxin 3. Nature. 2006;440:813–817. - PubMed
-
- Marszalek J.R., Lodish H.F. Docosahexaenoic acid, fatty acid-interacting proteins, and neuronal function: Breastmilk and fish are good for you. Annu. Rev. Cell Dev. Biol. 2005;21:633–657. - PubMed
-
- Ratnayake W.M., Galli C. Fat and fatty acid terminology, methods of analysis and fat digestion and metabolism: A background review paper. Ann. Nutr. Metab. 2009;55:8–43. - PubMed
-
- Nakamura M.T., Nara T.Y. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu. Rev. Nutr. 2004;24:345–376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

