High-throughput screening for genes that prevent excess DNA replication in human cells and for molecules that inhibit them
- PMID: 22503772
- PMCID: PMC4149752
- DOI: 10.1016/j.ymeth.2012.03.031
High-throughput screening for genes that prevent excess DNA replication in human cells and for molecules that inhibit them
Abstract
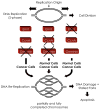
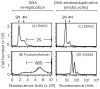
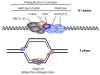
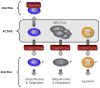
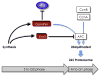
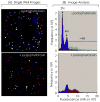
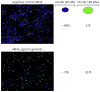
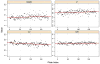
High-throughput screening (HTS) provides a rapid and comprehensive approach to identifying compounds that target specific biological processes as well as genes that are essential to those processes. Here we describe a HTS assay for small molecules that induce either DNA re-replication or endoreduplication (i.e. excess DNA replication) selectively in cells derived from human cancers. Such molecules will be useful not only to investigate cell division and differentiation, but they may provide a novel approach to cancer chemotherapy. Since induction of DNA re-replication results in apoptosis, compounds that selectively induce DNA re-replication in cancer cells without doing so in normal cells could kill cancers in vivo without preventing normal cell proliferation. Furthermore, the same HTS assay can be adapted to screen siRNA molecules to identify genes whose products restrict genome duplication to once per cell division. Some of these genes might regulate the formation of terminally differentiated polyploid cells during normal human development, whereas others will prevent DNA re-replication during each cell division. Based on previous studies, we anticipate that one or more of the latter genes will prove to be essential for proliferation of cancer cells but not for normal cells, since many cancer cells are deficient in mechanisms that maintain genome stability.
Copyright © 2012. Published by Elsevier Inc.
Figures

References
-
- Depamphilis ML, Bell SD. Genome Duplication. 1. Garland Science; London: 2010.
-
- Sivaprasad U, Dutta A, Bell SP. Assembly of pre-replication complexes. In: DePamphilis ML, editor. DNA Replication and Human Disease. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 63–88.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

