Overexpression of genes encoding glycolytic enzymes in Corynebacterium glutamicum enhances glucose metabolism and alanine production under oxygen deprivation conditions
- PMID: 22504802
- PMCID: PMC3370556
- DOI: 10.1128/AEM.07998-11
Overexpression of genes encoding glycolytic enzymes in Corynebacterium glutamicum enhances glucose metabolism and alanine production under oxygen deprivation conditions
Abstract
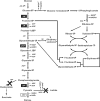
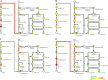
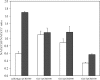
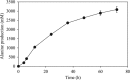
We previously reported that Corynebacterium glutamicum strain ΔldhAΔppc+alaD+gapA, overexpressing glyceraldehyde-3-phosphate dehydrogenase-encoding gapA, shows significantly improved glucose consumption and alanine formation under oxygen deprivation conditions (T. Jojima, M. Fujii, E. Mori, M. Inui, and H. Yukawa, Appl. Microbiol. Biotechnol. 87:159-165, 2010). In this study, we employ stepwise overexpression and chromosomal integration of a total of four genes encoding glycolytic enzymes (herein referred to as glycolytic genes) to demonstrate further successive improvements in C. glutamicum glucose metabolism under oxygen deprivation. In addition to gapA, overexpressing pyruvate kinase-encoding pyk and phosphofructokinase-encoding pfk enabled strain GLY2/pCRD500 to realize respective 13% and 20% improved rates of glucose consumption and alanine formation compared to GLY1/pCRD500. Subsequent overexpression of glucose-6-phosphate isomerase-encoding gpi in strain GLY3/pCRD500 further improved its glucose metabolism. Notably, both alanine productivity and yield increased after each overexpression step. After 48 h of incubation, GLY3/pCRD500 produced 2,430 mM alanine at a yield of 91.8%. This was 6.4-fold higher productivity than that of the wild-type strain. Intracellular metabolite analysis showed that gapA overexpression led to a decreased concentration of metabolites upstream of glyceraldehyde-3-phosphate dehydrogenase, suggesting that the overexpression resolved a bottleneck in glycolysis. Changing ratios of the extracellular metabolites by overexpression of glycolytic genes resulted in reduction of the intracellular NADH/NAD(+) ratio, which also plays an important role on the improvement of glucose consumption. Enhanced alanine dehydrogenase activity using a high-copy-number plasmid further accelerated the overall alanine productivity. Increase in glycolytic enzyme activities is a promising approach to make drastic progress in growth-arrested bioprocesses.
Figures


References
-
- Babul J, Clifton D, Kretschmer M, Fraenkel DG. 1993. Glucose metabolism in Escherichia coli and the effect of increased amount of aldolase. Biochemistry 32:4685–4692 - PubMed
-
- Davies SE, Brindle KM. 1992. Effects of overexpression of phosphofructokinase on glycolysis in the yeast Saccharomyces cerevisiae. Biochemistry 31:4729–4735 - PubMed
-
- Dominguez H, et al. 1998. Carbon-flux distribution in the central metabolic pathways of Corynebacterium glutamicum during growth on fructose. European journal of biochemistry / FEBS. 254:96–102 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

