High diversity and novel species of Pseudomonas aeruginosa bacteriophages
- PMID: 22504803
- PMCID: PMC3370533
- DOI: 10.1128/AEM.00065-12
High diversity and novel species of Pseudomonas aeruginosa bacteriophages
Abstract
The diversity of Pseudomonas aeruginosa bacteriophages was investigated using a collection of 68 phages isolated from Central Mexico. Most of the phages carried double-stranded DNA (dsDNA) genomes and were classified into 12 species. Comparison of the genomes of selected archetypal phages with extant sequences in GenBank resulted in the identification of six novel species. This finding increased the group diversity by ~30%. The great diversity of phage species could be related to the ubiquitous nature of P. aeruginosa.
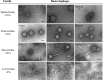
Figures

References
-
- Ackermann HW. 2007. 5500 phages examined in the electron microscope. Arch. Virol. 152:227–243 - PubMed
-
- Ackermann HW. 2006. Classification of bacteriophages, p 8–16 In Calendar RL, Abedon ST. (ed), The bacteriophages. Oxford University Press, Inc., New York, NY
-
- Ackermann HW, Cartier C, Slopek S, Vieu JF. 1988. Morphology of Pseudomonas aeruginosa typing phages of the Lindberg set. Ann. Inst. Pasteur Virol. 139:389–404 - PubMed
-
- Ackermann HW, et al. 1992. The species concept and its application to tailed phages. Arch. Virol. 124:69–82 - PubMed
-
- Ackermann HW, Eisenstark A. 1974. The present state of phage taxonomy. Intervirology 3:201–219 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

