The mechanism of γ-Secretase dysfunction in familial Alzheimer disease
- PMID: 22505025
- PMCID: PMC3364747
- DOI: 10.1038/emboj.2012.79
The mechanism of γ-Secretase dysfunction in familial Alzheimer disease
Abstract
The mechanisms by which mutations in the presenilins (PSEN) or the amyloid precursor protein (APP) genes cause familial Alzheimer disease (FAD) are controversial. FAD mutations increase the release of amyloid β (Aβ)42 relative to Aβ40 by an unknown, possibly gain-of-toxic-function, mechanism. However, many PSEN mutations paradoxically impair γ-secretase and 'loss-of-function' mechanisms have also been postulated. Here, we use kinetic studies to demonstrate that FAD mutations affect Aβ generation via three different mechanisms, resulting in qualitative changes in the Aβ profiles, which are not limited to Aβ42. Loss of ɛ-cleavage function is not generally observed among FAD mutants. On the other hand, γ-secretase inhibitors used in the clinic appear to block the initial ɛ-cleavage step, but unexpectedly affect more selectively Notch than APP processing, while modulators act as activators of the carboxypeptidase-like (γ) activity. Overall, we provide a coherent explanation for the effect of different FAD mutations, demonstrating the importance of qualitative rather than quantitative changes in the Aβ products, and suggest fundamental improvements for current drug development efforts.
Conflict of interest statement
Harrie Gijsen is an employee of Janssen Pharmaceutica. Bart de Strooper is a consultant for Janssen Pharmaceutica, Envivo Pharmaceutics and Remynd and is supported by research grants from Janssen Pharmaceutics.
Figures
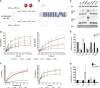
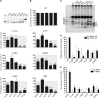
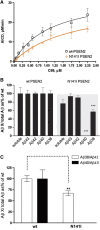

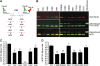
Comment in
-
Shifting a complex debate on γ-secretase cleavage and Alzheimer's disease.EMBO J. 2012 May 16;31(10):2237-9. doi: 10.1038/emboj.2012.111. Epub 2012 Apr 13. EMBO J. 2012. PMID: 22505028 Free PMC article.
References
-
- Annaert WG, Esselens C, Baert V, Boeve C, Snellings G, Cupers P, Craessaerts K, De Strooper B (2001) Interaction with telencephalin and the amyloid precursor protein predicts a ring structure for presenilins. Neuron 32: 579–589 - PubMed
-
- Annaert WG, Levesque L, Craessaerts K, Dierinck I, Snellings G, Westaway D, George-Hyslop PS, Cordell B, Fraser P, De Strooper B (1999) Presenilin 1 controls gamma-secretase processing of amyloid precursor protein in pre-golgi compartments of hippocampal neurons. J Cell Biol 147: 277–294 - PMC - PubMed
-
- Bentahir M, Nyabi O, Verhamme J, Tolia A, Horre K, Wiltfang J, Esselmann H, De Strooper B (2006) Presenilin clinical mutations can affect gamma-secretase activity by different mechanisms. J Neurochem 96: 732–742 - PubMed
-
- Bergmans BA, De Strooper B (2010) Gamma-secretases: from cell biology to therapeutic strategies. Lancet Neurol 9: 2215–226 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

