JLigand: a graphical tool for the CCP4 template-restraint library
- PMID: 22505263
- PMCID: PMC3322602
- DOI: 10.1107/S090744491200251X
JLigand: a graphical tool for the CCP4 template-restraint library
Abstract
Biological macromolecules are polymers and therefore the restraints for macromolecular refinement can be subdivided into two sets: restraints that are applied to atoms that all belong to the same monomer and restraints that are associated with the covalent bonds between monomers. The CCP4 template-restraint library contains three types of data entries defining template restraints: descriptions of monomers and their modifications, both used for intramonomer restraints, and descriptions of links for intermonomer restraints. The library provides generic descriptions of modifications and links for protein, DNA and RNA chains, and for some post-translational modifications including glycosylation. Structure-specific template restraints can be defined in a user's additional restraint library. Here, JLigand, a new CCP4 graphical interface to LibCheck and REFMAC that has been developed to manage the user's library and generate new monomer entries is described, as well as new entries for links and associated modifications.
Figures
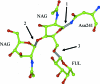


References
-
- Bernstein, F. C., Koetzle, T. F., Williams, G. J., Meyer, E. F. Jr, Brice, M. D., Rodgers, J. R., Kennard, O., Shimanouchi, T. & Tasumi, M. (1977). J. Mol. Biol. 112, 535–542. - PubMed
-
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P., Sharff, A., Smart, O. S., Vonrhein, C. & Womack, T. O. (2011). BUSTER Cambridge: Global Phasing Ltd.
-
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
Publication types
MeSH terms
Substances
Grants and funding
- MC_UP_A025_1012/MRC_/Medical Research Council/United Kingdom
- BB/F020228/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 064405/Z/01/A/WT_/Wellcome Trust/United Kingdom
- BB/F001134/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

